Selection and Prioritization of Candidate Drug Targets for Amyotrophic Lateral Sclerosis Through a Meta-Analysis Approach
- PMID: 28236105
- PMCID: PMC5359376
- DOI: 10.1007/s12031-017-0898-9
Selection and Prioritization of Candidate Drug Targets for Amyotrophic Lateral Sclerosis Through a Meta-Analysis Approach
Abstract
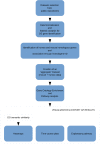
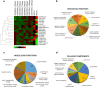
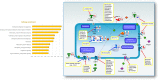
Amyotrophic lateral sclerosis (ALS) is a progressive and incurable neurodegenerative disease. Although several compounds have shown promising results in preclinical studies, their translation into clinical trials has failed. This clinical failure is likely due to the inadequacy of the animal models that do not sufficiently reflect the human disease. Therefore, it is important to optimize drug target selection by identifying those that overlap in human and mouse pathology. We have recently characterized the transcriptional profiles of motor cortex samples from sporadic ALS (SALS) patients and differentiated these into two subgroups based on differentially expressed genes, which encode 70 potential therapeutic targets. To prioritize drug target selection, we investigated their degree of conservation in superoxide dismutase 1 (SOD1) G93A transgenic mice, the most widely used ALS animal model. Interspecies comparison of our human expression data with those of eight different SOD1G93A datasets present in public repositories revealed the presence of commonly deregulated targets and related biological processes. Moreover, deregulated expression of the majority of our candidate targets occurred at the onset of the disease, offering the possibility to use them for an early and more effective diagnosis and therapy. In addition to highlighting the existence of common key drivers in human and mouse pathology, our study represents the basis for a rational preclinical drug development.
Keywords: ALS; Meta-analysis; SOD1G93A mouse model; Transcriptomics.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Similar articles
-
Therapeutic immunization with a glatiramer acetate derivative does not alter survival in G93A and G37R SOD1 mouse models of familial ALS.Neurobiol Dis. 2007 Apr;26(1):146-52. doi: 10.1016/j.nbd.2006.12.013. Epub 2006 Dec 30. Neurobiol Dis. 2007. PMID: 17276077
-
Guanabenz delays the onset of disease symptoms, extends lifespan, improves motor performance and attenuates motor neuron loss in the SOD1 G93A mouse model of amyotrophic lateral sclerosis.Neuroscience. 2014 Sep 26;277:132-8. doi: 10.1016/j.neuroscience.2014.03.047. Epub 2014 Mar 31. Neuroscience. 2014. PMID: 24699224
-
Transcriptomic indices of fast and slow disease progression in two mouse models of amyotrophic lateral sclerosis.Brain. 2013 Nov;136(Pt 11):3305-32. doi: 10.1093/brain/awt250. Epub 2013 Sep 24. Brain. 2013. PMID: 24065725
-
Transcriptional analysis reveals distinct subtypes in amyotrophic lateral sclerosis: implications for personalized therapy.Future Med Chem. 2015;7(10):1335-59. doi: 10.4155/fmc.15.60. Future Med Chem. 2015. PMID: 26144267 Review.
-
Motor neuron disease: a chemical perspective.J Med Chem. 2014 Aug 14;57(15):6316-31. doi: 10.1021/jm5001584. Epub 2014 Apr 17. J Med Chem. 2014. PMID: 24694032 Review.
Cited by
-
Taxonomy Meets Neurology, the Case of Amyotrophic Lateral Sclerosis.Front Neurosci. 2018 Sep 26;12:673. doi: 10.3389/fnins.2018.00673. eCollection 2018. Front Neurosci. 2018. PMID: 30319346 Free PMC article.
-
From Multi-Omics Approaches to Precision Medicine in Amyotrophic Lateral Sclerosis.Front Neurosci. 2020 Oct 30;14:577755. doi: 10.3389/fnins.2020.577755. eCollection 2020. Front Neurosci. 2020. PMID: 33192262 Free PMC article. Review.
-
A Diagnostic Gene-Expression Signature in Fibroblasts of Amyotrophic Lateral Sclerosis.Cells. 2023 Jul 18;12(14):1884. doi: 10.3390/cells12141884. Cells. 2023. PMID: 37508548 Free PMC article.
-
Splicing Players Are Differently Expressed in Sporadic Amyotrophic Lateral Sclerosis Molecular Clusters and Brain Regions.Cells. 2020 Jan 8;9(1):159. doi: 10.3390/cells9010159. Cells. 2020. PMID: 31936368 Free PMC article.
-
CXCR2 Is Deregulated in ALS Spinal Cord and Its Activation Triggers Apoptosis in Motor Neuron-Like Cells Overexpressing hSOD1-G93A.Cells. 2023 Jul 9;12(14):1813. doi: 10.3390/cells12141813. Cells. 2023. PMID: 37508478 Free PMC article.
References
-
- Achilli F, Boyle S, Kieran D, Chia R, Hafezparast M, Martin JE, Schiavo G, Greensmith L, Bickmore W, Fisher EM. The SOD1 transgene in the G93A mouse model of amyotrophic lateral sclerosis lies on distal mouse chromosome 12. Amyotrophic lateral sclerosis and other motor neuron disorders: official publication of the World Federation of Neurology, Research Group on Motor Neuron Diseases. 2005;6(2):111–114. doi: 10.1080/14660820510035351. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

