A calmodulin-like protein suppresses RNA silencing and promotes geminivirus infection by degrading SGS3 via the autophagy pathway in Nicotiana benthamiana
- PMID: 28212430
- PMCID: PMC5333915
- DOI: 10.1371/journal.ppat.1006213
A calmodulin-like protein suppresses RNA silencing and promotes geminivirus infection by degrading SGS3 via the autophagy pathway in Nicotiana benthamiana
Abstract
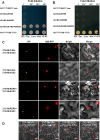
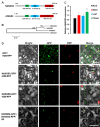
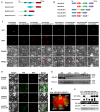
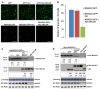
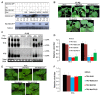
A recently characterized calmodulin-like protein is an endogenous RNA silencing suppressor that suppresses sense-RNA induced post-transcriptional gene silencing (S-PTGS) and enhances virus infection, but the mechanism underlying calmodulin-like protein-mediated S-PTGS suppression is obscure. Here, we show that a calmodulin-like protein from Nicotiana benthamiana (NbCaM) interacts with Suppressor of Gene Silencing 3 (NbSGS3). Deletion analyses showed that domains essential for the interaction between NbSGS3 and NbCaM are also required for the subcellular localization of NbSGS3 and NbCaM suppressor activity. Overexpression of NbCaM reduced the number of NbSGS3-associated granules by degrading NbSGS3 protein accumulation in the cytoplasm. This NbCaM-mediated NbSGS3 degradation was sensitive to the autophagy inhibitors 3-methyladenine and E64d, and was compromised when key autophagy genes of the phosphatidylinositol 3-kinase (PI3K) complex were knocked down. Meanwhile, silencing of key autophagy genes within the PI3K complex inhibited geminivirus infection. Taken together these data suggest that NbCaM acts as a suppressor of RNA silencing by degrading NbSGS3 through the autophagy pathway.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
SGS3 Cooperates with RDR6 in Triggering Geminivirus-Induced Gene Silencing and in Suppressing Geminivirus Infection in Nicotiana Benthamiana.Viruses. 2017 Sep 4;9(9):247. doi: 10.3390/v9090247. Viruses. 2017. PMID: 28869553 Free PMC article.
-
Suppression of RNA silencing by a plant DNA virus satellite requires a host calmodulin-like protein to repress RDR6 expression.PLoS Pathog. 2014 Feb 6;10(2):e1003921. doi: 10.1371/journal.ppat.1003921. eCollection 2014 Feb. PLoS Pathog. 2014. PMID: 24516387 Free PMC article.
-
Geminivirus AL2 protein induces expression of, and interacts with, a calmodulin-like gene, an endogenous regulator of gene silencing.Virology. 2014 Jul;460-461:108-18. doi: 10.1016/j.virol.2014.04.034. Epub 2014 May 29. Virology. 2014. PMID: 25010276
-
Silencing suppression by geminivirus proteins.Virology. 2006 Jan 5;344(1):158-68. doi: 10.1016/j.virol.2005.09.041. Virology. 2006. PMID: 16364747 Review.
-
Geminivirus C4: Interplaying with Receptor-like Kinases.Trends Plant Sci. 2018 Dec;23(12):1044-1046. doi: 10.1016/j.tplants.2018.09.003. Epub 2018 Sep 29. Trends Plant Sci. 2018. PMID: 30279072 Review.
Cited by
-
An asymptomatic geminivirus activates autophagy and enhances plant defenses against diverse pathogens.Stress Biol. 2024 Oct 8;4(1):42. doi: 10.1007/s44154-024-00176-8. Stress Biol. 2024. PMID: 39377848 Free PMC article.
-
Stress-related biomolecular condensates in plants.Plant Cell. 2023 Sep 1;35(9):3187-3204. doi: 10.1093/plcell/koad127. Plant Cell. 2023. PMID: 37162152 Free PMC article.
-
Dynamic Subcellular Localization, Accumulation, and Interactions of Proteins From Tomato Yellow Leaf Curl China Virus and Its Associated Betasatellite.Front Plant Sci. 2020 Jun 16;11:840. doi: 10.3389/fpls.2020.00840. eCollection 2020. Front Plant Sci. 2020. PMID: 32612626 Free PMC article.
-
Autophagy-virus interplay in plants: from antiviral recognition to proviral manipulation.Mol Plant Pathol. 2019 Sep;20(9):1211-1216. doi: 10.1111/mpp.12852. Epub 2019 Aug 9. Mol Plant Pathol. 2019. PMID: 31397085 Free PMC article. Review.
-
The nonstructural protein NSs encoded by tomato zonate spot virus suppresses RNA silencing by interacting with NbSGS3.Mol Plant Pathol. 2022 May;23(5):707-719. doi: 10.1111/mpp.13192. Epub 2022 Feb 20. Mol Plant Pathol. 2022. PMID: 35184365 Free PMC article.
References
-
- Bologna NG, Voinnet O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol. 2014; 28: 1–31. - PubMed
-
- Mourrain P, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000; 101: 533–542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

