The VDAC2-BAK axis regulates peroxisomal membrane permeability
- PMID: 28174205
- PMCID: PMC5350511
- DOI: 10.1083/jcb.201605002
The VDAC2-BAK axis regulates peroxisomal membrane permeability
Abstract
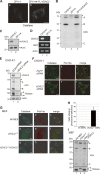
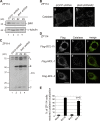
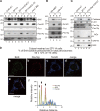
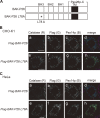
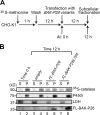
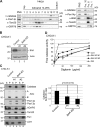
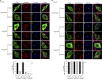
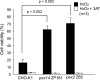
Peroxisomal biogenesis disorders (PBDs) are fatal genetic diseases consisting of 14 complementation groups (CGs). We previously isolated a peroxisome-deficient Chinese hamster ovary cell mutant, ZP114, which belongs to none of these CGs. Using a functional screening strategy, VDAC2 was identified as rescuing the peroxisomal deficiency of ZP114 where VDAC2 expression was not detected. Interestingly, knockdown of BAK or overexpression of the BAK inhibitors BCL-XL and MCL-1 restored peroxisomal biogenesis in ZP114 cells. Although VDAC2 is not localized to the peroxisome, loss of VDAC2 shifts the localization of BAK from mitochondria to peroxisomes, resulting in peroxisomal deficiency. Introduction of peroxisome-targeted BAK harboring the Pex26p transmembrane region into wild-type cells resulted in the release of peroxisomal matrix proteins to cytosol. Moreover, overexpression of BAK activators PUMA and BIM permeabilized peroxisomes in a BAK-dependent manner. Collectively, these findings suggest that BAK plays a role in peroxisomal permeability, similar to mitochondrial outer membrane permeabilization.
© 2017 Hosoi et al.
Figures
Similar articles
-
The peroxisomes strike BAK: Regulation of peroxisome integrity by the Bcl-2 family.J Cell Biol. 2017 Mar 6;216(3):547-549. doi: 10.1083/jcb.201701187. Epub 2017 Feb 13. J Cell Biol. 2017. PMID: 28193702 Free PMC article.
-
BAK regulates catalase release from peroxisomes.Mol Cell Oncol. 2017 Mar 17;4(3):e1306610. doi: 10.1080/23723556.2017.1306610. eCollection 2017. Mol Cell Oncol. 2017. PMID: 28616584 Free PMC article.
-
Bax targets mitochondria by distinct mechanisms before or during apoptotic cell death: a requirement for VDAC2 or Bak for efficient Bax apoptotic function.Cell Death Differ. 2014 Dec;21(12):1925-35. doi: 10.1038/cdd.2014.119. Epub 2014 Aug 22. Cell Death Differ. 2014. PMID: 25146925 Free PMC article.
-
Cell Death or Survival Against Oxidative Stress.Subcell Biochem. 2018;89:463-471. doi: 10.1007/978-981-13-2233-4_20. Subcell Biochem. 2018. PMID: 30378036 Review.
-
Peroxisome biogenesis and human peroxisome-deficiency disorders.Proc Jpn Acad Ser B Phys Biol Sci. 2016;92(10):463-477. doi: 10.2183/pjab.92.463. Proc Jpn Acad Ser B Phys Biol Sci. 2016. PMID: 27941306 Free PMC article. Review.
Cited by
-
Exposure to the plasticizer dibutyl phthalate causes oxidative stress and neurotoxicity in brain tissue.Environ Sci Pollut Res Int. 2024 Mar;31(14):21399-21414. doi: 10.1007/s11356-024-32604-7. Epub 2024 Feb 23. Environ Sci Pollut Res Int. 2024. PMID: 38393557
-
Human Cytomegalovirus vMIA Inhibits MAVS Oligomerization at Peroxisomes in an MFF-Dependent Manner.Front Cell Dev Biol. 2022 Apr 4;10:871977. doi: 10.3389/fcell.2022.871977. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35445031 Free PMC article.
-
Androgens Modulate Bcl-2 Agonist of Cell Death (BAD) Expression and Function in Breast Cancer Cells.Int J Mol Sci. 2023 Aug 30;24(17):13464. doi: 10.3390/ijms241713464. Int J Mol Sci. 2023. PMID: 37686282 Free PMC article.
-
Mitochondrial and Organellar Crosstalk in Parkinson's Disease.ASN Neuro. 2021 Jan-Dec;13:17590914211028364. doi: 10.1177/17590914211028364. ASN Neuro. 2021. PMID: 34304614 Free PMC article.
-
Predicting the targeting of tail-anchored proteins to subcellular compartments in mammalian cells.J Cell Sci. 2017 May 1;130(9):1675-1687. doi: 10.1242/jcs.200204. Epub 2017 Mar 21. J Cell Sci. 2017. PMID: 28325759 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

