Hepatitis B Virus-Encoded MicroRNA Controls Viral Replication
- PMID: 28148795
- PMCID: PMC5411615
- DOI: 10.1128/JVI.01919-16
Hepatitis B Virus-Encoded MicroRNA Controls Viral Replication
Abstract
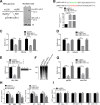
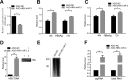
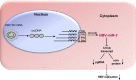
MicroRNAs (miRNAs) are a class of small, single-stranded, noncoding, functional RNAs. Hepatitis B virus (HBV) is an enveloped DNA virus with virions and subviral forms of particles that lack a core. It was not known whether HBV encodes miRNAs. Here, we identified an HBV-encoded miRNA (called HBV-miR-3) by deep sequencing and Northern blotting. HBV-miR-3 is located at nucleotides (nt) 373 to 393 of the HBV genome and was generated from 3.5-kb, 2.4-kb, and 2.1-kb HBV in a classic miRNA biogenesis (Drosha-Dicer-dependent) manner. HBV-miR-3 was highly expressed in hepatoma cell lines with an integrated HBV genome and HBV+ hepatoma tumors. In patients with HBV infection, HBV-miR-3 was released into the circulation by exosomes and HBV virions, and HBV-miR-3 expression had a positive correlation with HBV titers in the sera of patients in the acute phase of HBV infection. More interestingly, we found that HBV-miR-3 represses HBsAg, HBeAg, and replication of HBV. HBV-miR-3 targets the unique site of the HBV 3.5-kb transcript to specifically reduce HBc protein expression, levels of pregenomic RNA (pgRNA), and HBV replication intermediate (HBV-RI) generation but does not affect the HBV DNA polymerase level, thus suppressing HBV virion production (replication). This may explain the low levels of HBV virion generation with abundant subviral particles lacking core during HBV replication, which may contribute to the development of persistent infection in patients. Taken together, our findings shed light on novel mechanisms by which HBV-encoded miRNA controls the process of self-replication by regulating HBV transcript during infection.IMPORTANCE Hepatitis B is a liver infection caused by the hepatitis B virus (HBV) that can become a long-term, chronic infection and lead to cirrhosis or liver cancer. HBV is a small DNA virus that belongs to the hepadnavirus family, with virions and subviral forms of particles that lack a core. MicroRNA (miRNA), a small (∼22-nt) noncoding RNA, was recently found to be an important regulator of gene expression. We found that HBV encodes miRNA (HBV-miR-3). More importantly, we revealed that HBV-miR-3 targets its transcripts to attenuate HBV replication. This may contribute to explaining how HBV infection leads to mild damage in liver cells and the subsequent establishment/maintenance of persistent infection. Our findings highlight a mechanism by which HBV-encoded miRNA controls the process of self-replication by regulating the virus itself during infection and might provide new biomarkers for diagnosis and treatment of hepatitis B.
Keywords: HBc; hepatitis B virus; miRNA; pgRNA.
Copyright © 2017 American Society for Microbiology.
Figures




Similar articles
-
miR-200c targets nuclear factor IA to suppress HBV replication and gene expression via repressing HBV Enhancer I activity.Biomed Pharmacother. 2018 Mar;99:774-780. doi: 10.1016/j.biopha.2018.01.141. Epub 2018 Feb 20. Biomed Pharmacother. 2018. PMID: 29710475
-
Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210.Antiviral Res. 2010 Nov;88(2):169-75. doi: 10.1016/j.antiviral.2010.08.008. Epub 2010 Aug 20. Antiviral Res. 2010. PMID: 20728471
-
Lost Small Envelope Protein Expression from Naturally Occurring PreS1 Deletion Mutants of Hepatitis B Virus Is Often Accompanied by Increased HBx and Core Protein Expression as Well as Genome Replication.J Virol. 2021 Jun 24;95(14):e0066021. doi: 10.1128/JVI.00660-21. Epub 2021 Jun 24. J Virol. 2021. PMID: 33910956 Free PMC article.
-
Intracellular Trafficking of HBV Particles.Cells. 2020 Sep 2;9(9):2023. doi: 10.3390/cells9092023. Cells. 2020. PMID: 32887393 Free PMC article. Review.
-
Role of microRNAs in hepatitis B virus replication and pathogenesis.Biochim Biophys Acta. 2011 Nov-Dec;1809(11-12):678-85. doi: 10.1016/j.bbagrm.2011.04.008. Epub 2011 May 4. Biochim Biophys Acta. 2011. PMID: 21565290 Review.
Cited by
-
Why Cells and Viruses Cannot Survive without an ESCRT.Cells. 2021 Feb 24;10(3):483. doi: 10.3390/cells10030483. Cells. 2021. PMID: 33668191 Free PMC article. Review.
-
Identification of a Novel HBV Encoded miRNA Using Next Generation Sequencing.Viruses. 2022 Jun 5;14(6):1223. doi: 10.3390/v14061223. Viruses. 2022. PMID: 35746694 Free PMC article.
-
Circulating microRNAs as non-invasive biomarkers for hepatitis B virus liver fibrosis.World J Gastroenterol. 2020 Mar 21;26(11):1113-1127. doi: 10.3748/wjg.v26.i11.1113. World J Gastroenterol. 2020. PMID: 32231417 Free PMC article. Review.
-
Clinical implications of exosome-derived noncoding RNAs in liver.Lab Invest. 2022 May;102(5):464-473. doi: 10.1038/s41374-021-00723-1. Epub 2022 Jan 10. Lab Invest. 2022. PMID: 35013531 Review.
-
Intracellular bacteria engage a STING-TBK1-MVB12b pathway to enable paracrine cGAS-STING signalling.Nat Microbiol. 2019 Apr;4(4):701-713. doi: 10.1038/s41564-019-0367-z. Epub 2019 Feb 25. Nat Microbiol. 2019. PMID: 30804548 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

