Feedback control of AHR signalling regulates intestinal immunity
- PMID: 28146477
- PMCID: PMC5302159
- DOI: 10.1038/nature21080
Feedback control of AHR signalling regulates intestinal immunity
Abstract
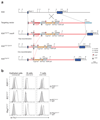
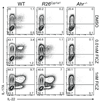
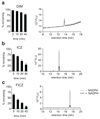
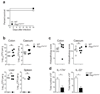
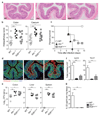
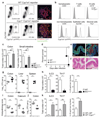
The aryl hydrocarbon receptor (AHR) recognizes xenobiotics as well as natural compounds such as tryptophan metabolites, dietary components and microbiota-derived factors, and it is important for maintenance of homeostasis at mucosal surfaces. AHR activation induces cytochrome P4501 (CYP1) enzymes, which oxygenate AHR ligands, leading to their metabolic clearance and detoxification. Thus, CYP1 enzymes have an important feedback role that curtails the duration of AHR signalling, but it remains unclear whether they also regulate AHR ligand availability in vivo. Here we show that dysregulated expression of Cyp1a1 in mice depletes the reservoir of natural AHR ligands, generating a quasi AHR-deficient state. Constitutive expression of Cyp1a1 throughout the body or restricted specifically to intestinal epithelial cells resulted in loss of AHR-dependent type 3 innate lymphoid cells and T helper 17 cells and increased susceptibility to enteric infection. The deleterious effects of excessive AHR ligand degradation on intestinal immune functions could be counter-balanced by increasing the intake of AHR ligands in the diet. Thus, our data indicate that intestinal epithelial cells serve as gatekeepers for the supply of AHR ligands to the host and emphasize the importance of feedback control in modulating AHR pathway activation.
Conflict of interest statement
Author information: The authors declare no competing financial interests.
Figures

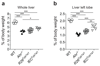


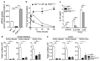

Comment in
-
Mucosal immunology: Rationing AHR ligands.Nat Rev Immunol. 2017 Mar;17(3):146-147. doi: 10.1038/nri.2017.8. Epub 2017 Feb 13. Nat Rev Immunol. 2017. PMID: 28192416 No abstract available.
Similar articles
-
Cytochrome P4501-inhibiting chemicals amplify aryl hydrocarbon receptor activation and IL-22 production in T helper 17 cells.Biochem Pharmacol. 2018 May;151:47-58. doi: 10.1016/j.bcp.2018.02.031. Epub 2018 Mar 1. Biochem Pharmacol. 2018. PMID: 29501585
-
How the AHR Became Important in Intestinal Homeostasis-A Diurnal FICZ/AHR/CYP1A1 Feedback Controls Both Immunity and Immunopathology.Int J Mol Sci. 2020 Aug 8;21(16):5681. doi: 10.3390/ijms21165681. Int J Mol Sci. 2020. PMID: 32784381 Free PMC article. Review.
-
Aryl hydrocarbon receptor promotes RORγt⁺ group 3 ILCs and controls intestinal immunity and inflammation.Semin Immunopathol. 2013 Nov;35(6):657-70. doi: 10.1007/s00281-013-0393-5. Epub 2013 Aug 23. Semin Immunopathol. 2013. PMID: 23975386 Free PMC article. Review.
-
Norisoboldine, an isoquinoline alkaloid, acts as an aryl hydrocarbon receptor ligand to induce intestinal Treg cells and thereby attenuate arthritis.Int J Biochem Cell Biol. 2016 Jun;75:63-73. doi: 10.1016/j.biocel.2016.03.014. Epub 2016 Mar 28. Int J Biochem Cell Biol. 2016. PMID: 27032495
-
Aryl Hydrocarbon Receptor Signaling Cell Intrinsically Inhibits Intestinal Group 2 Innate Lymphoid Cell Function.Immunity. 2018 Nov 20;49(5):915-928.e5. doi: 10.1016/j.immuni.2018.09.015. Epub 2018 Nov 13. Immunity. 2018. PMID: 30446384 Free PMC article.
Cited by
-
Intestinal microbiota-derived tryptophan metabolites are predictive of Ah receptor activity.Gut Microbes. 2020 Nov 9;12(1):1-24. doi: 10.1080/19490976.2020.1788899. Gut Microbes. 2020. PMID: 32783770 Free PMC article.
-
AhR ligands from LGG metabolites promote piglet intestinal ILC3 activation and IL-22 secretion to inhibit PEDV infection.J Virol. 2024 Aug 20;98(8):e0103924. doi: 10.1128/jvi.01039-24. Epub 2024 Jul 16. J Virol. 2024. PMID: 39012142 Free PMC article.
-
T Cell Responses to the Microbiota.Annu Rev Immunol. 2022 Apr 26;40:559-587. doi: 10.1146/annurev-immunol-101320-011829. Epub 2022 Feb 3. Annu Rev Immunol. 2022. PMID: 35113732 Free PMC article. Review.
-
Food Chemicals Disrupt Human Gut Microbiota Activity And Impact Intestinal Homeostasis As Revealed By In Vitro Systems.Sci Rep. 2018 Jul 20;8(1):11006. doi: 10.1038/s41598-018-29376-9. Sci Rep. 2018. PMID: 30030472 Free PMC article.
-
Fecal Microbiota Transplantation Ameliorates Experimentally Induced Colitis in Mice by Upregulating AhR.Front Microbiol. 2018 Aug 24;9:1921. doi: 10.3389/fmicb.2018.01921. eCollection 2018. Front Microbiol. 2018. PMID: 30197631 Free PMC article.
References
-
- Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annual review of pharmacology and toxicology. 2003;43:309–334. - PubMed
-
- Zelante T, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. - PubMed
-
- McMillan BJ, Bradfield CA. The aryl hydrocarbon receptor sans xenobiotics: endogenous function in genetic model systems. Mol Pharmacol. 2007;72:487–498. - PubMed
-
- Moura-Alves P, et al. AhR sensing of bacterial pigments regulates antibacterial defence. Nature. 2014;512:387–392. - PubMed
-
- Schmidt JV, Bradfield CA. Ah receptor signaling pathways. Annual review of cell and developmental biology. 1996;12:55–89. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

