The isolation of morphologically intact and biologically active extracellular vesicles from the secretome of cancer-associated adipose tissue
- PMID: 28146372
- PMCID: PMC5351718
- DOI: 10.1080/19336918.2017.1279784
The isolation of morphologically intact and biologically active extracellular vesicles from the secretome of cancer-associated adipose tissue
Abstract
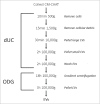
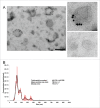
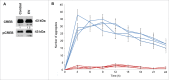
Breast cancer cells closely interact with different cell types of the surrounding adipose tissue to favor invasive growth and metastasis. Extracellular vesicles (EVs) are nanometer-sized vesicles secreted by different cell types that shuttle proteins and nucleic acids to establish cell-cell communication. To study the role of EVs released by cancer-associated adipose tissue in breast cancer progression and metastasis a standardized EV isolation protocol that obtains pure EVs and maintains their functional characteristics is required. We implemented differential ultracentrifugation as a pre-enrichment step followed by OptiPrep density gradient centrifugation (dUC-ODG) to isolate EVs from the conditioned medium of cancer-associated adipose tissue. A combination of immune-electron microscopy, nanoparticle tracking analysis (NTA) and Western blot analysis identified EVs that are enriched in flotillin-1, CD9 and CD63, and sized between 20 and 200 nm with a density of 1.076-1.125 g/ml. The lack of protein aggregates and cell organelle proteins confirmed the purity of the EV preparations. Next, we evaluated whether dUC-ODG isolated EVs are functionally active. ZR75.1 breast cancer cells treated with cancer-associated adipose tissue-secreted EVs from breast cancer patients showed an increased phosphorylation of CREB. MCF-7 breast cancer cells treated with adipose tissue-derived EVs exhibited a stronger propensity to form cellular aggregates. In conclusion, dUC-ODG purifies EVs from conditioned medium of cancer-associated adipose tissue, and these EVs are morphologically intact and biologically active.
Keywords: aggregation; breast cancer; characterization; exosomes; function; isolation; proliferation.
Figures

Similar articles
-
Isolation and Characterization of Functionally Active Extracellular Vesicles from Culture Medium Conditioned by Bovine Embryos In Vitro.Int J Mol Sci. 2018 Dec 21;20(1):38. doi: 10.3390/ijms20010038. Int J Mol Sci. 2018. PMID: 30577682 Free PMC article.
-
The Separation and Characterization of Extracellular Vesicles from Medium Conditioned by Bovine Embryos.Int J Mol Sci. 2020 Apr 22;21(8):2942. doi: 10.3390/ijms21082942. Int J Mol Sci. 2020. PMID: 32331414 Free PMC article.
-
Serum-derived extracellular vesicles from breast cancer patients contribute to differential regulation of T-cell-mediated immune-escape mechanisms in breast cancer subtypes.Front Immunol. 2023 Jun 22;14:1204224. doi: 10.3389/fimmu.2023.1204224. eCollection 2023. Front Immunol. 2023. PMID: 37441083 Free PMC article.
-
Subpopulations of extracellular vesicles and their therapeutic potential.Mol Aspects Med. 2018 Apr;60:1-14. doi: 10.1016/j.mam.2018.02.002. Epub 2018 Feb 16. Mol Aspects Med. 2018. PMID: 29432782 Review.
-
The Methods of Choice for Extracellular Vesicles (EVs) Characterization.Int J Mol Sci. 2017 May 29;18(6):1153. doi: 10.3390/ijms18061153. Int J Mol Sci. 2017. PMID: 28555055 Free PMC article. Review.
Cited by
-
Influence of species and processing parameters on recovery and content of brain tissue-derived extracellular vesicles.J Extracell Vesicles. 2020 Jun 30;9(1):1785746. doi: 10.1080/20013078.2020.1785746. J Extracell Vesicles. 2020. PMID: 32944174 Free PMC article.
-
Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches.J Extracell Vesicles. 2024 Feb;13(2):e12404. doi: 10.1002/jev2.12404. J Extracell Vesicles. 2024. PMID: 38326288 Free PMC article.
-
Prospects and challenges of tissue-derived extracellular vesicles.Mol Ther. 2024 Sep 4;32(9):2950-2978. doi: 10.1016/j.ymthe.2024.06.025. Epub 2024 Jun 22. Mol Ther. 2024. PMID: 38910325 Free PMC article. Review.
-
Hitting the Bullseye: Are extracellular vesicles on target?J Extracell Vesicles. 2020 Nov;10(1):e12032. doi: 10.1002/jev2.12032. Epub 2020 Nov 29. J Extracell Vesicles. 2020. PMID: 33708359 Free PMC article. No abstract available.
-
Proteomic profiling of tumour tissue-derived extracellular vesicles in colon cancer.J Extracell Biol. 2024 Feb 6;3(2):e127. doi: 10.1002/jex2.127. eCollection 2024 Feb. J Extracell Biol. 2024. PMID: 38939898 Free PMC article.
References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144(5):646-74; PMID:21376230; http://dx.doi.org/10.1016/j.cell.2011.02.013 - DOI - PubMed
-
- Lapeire L, Denys H, Cocquyt V, De Wever O. When fat becomes an ally of the enemy: adipose tissue as collaborator in human breast cancer. Horm Mol Biol Clin Investig 2015; 23(1):21-38; PMID:26154196; http://dx.doi.org/10.1515/hmbci-2015-0018 - DOI - PubMed
-
- Lapeire L, Hendrix A, Lambein K, Van Bockstal M, Braems G, Van Den Broecke R, Limame R, Mestdagh P, Vandesompele J, Vanhove C, et al.. Cancer-associated adipose tissue promotes breast cancer progression by paracrine oncostatin M and Jak/STAT3 signaling. Cancer Res 2014; 74(23):6806-19; PMID:25252914; http://dx.doi.org/10.1158/0008-5472.CAN-14-0160 - DOI - PubMed
-
- Dirat B, Bochet L, Dabek M, Daviaud D, Dauvillier S, Majed B, Wang YY, Meulle A, Salles B, Le Gonidec S, et al.. Cancer-associated adipocytes exhibit an activated phenotype and contribute to breast cancer invasion. Cancer Res 2011; 71(7):2455-65; PMID:21459803; http://dx.doi.org/10.1158/0008-5472.CAN-10-3323 - DOI - PubMed
-
- Hendrix A, Hume AN. Exosome signaling in mammary gland development and cancer. Int J Dev Biol 2011; 55(7–9):879-87; PMID:22161843; http://dx.doi.org/10.1387/ijdb.113391ah - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
