An Argonaute phosphorylation cycle promotes microRNA-mediated silencing
- PMID: 28114302
- PMCID: PMC5302127
- DOI: 10.1038/nature21025
An Argonaute phosphorylation cycle promotes microRNA-mediated silencing
Abstract
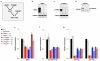
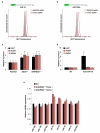
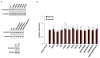
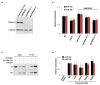
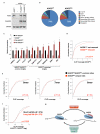
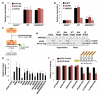
MicroRNAs (miRNAs) perform critical functions in normal physiology and disease by associating with Argonaute proteins and downregulating partially complementary messenger RNAs (mRNAs). Here we use clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (Cas9) genome-wide loss-of-function screening coupled with a fluorescent reporter of miRNA activity in human cells to identify new regulators of the miRNA pathway. By using iterative rounds of screening, we reveal a novel mechanism whereby target engagement by Argonaute 2 (AGO2) triggers its hierarchical, multi-site phosphorylation by CSNK1A1 on a set of highly conserved residues (S824-S834), followed by rapid dephosphorylation by the ANKRD52-PPP6C phosphatase complex. Although genetic and biochemical studies demonstrate that AGO2 phosphorylation on these residues inhibits target mRNA binding, inactivation of this phosphorylation cycle globally impairs miRNA-mediated silencing. Analysis of the transcriptome-wide binding profile of non-phosphorylatable AGO2 reveals a pronounced expansion of the target repertoire bound at steady-state, effectively reducing the active pool of AGO2 on a per-target basis. These findings support a model in which an AGO2 phosphorylation cycle stimulated by target engagement regulates miRNA:target interactions to maintain the global efficiency of miRNA-mediated silencing.
Figures

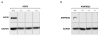






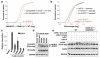

Similar articles
-
Target binding triggers hierarchical phosphorylation of human Argonaute-2 to promote target release.Elife. 2022 May 31;11:e76908. doi: 10.7554/eLife.76908. Elife. 2022. PMID: 35638597 Free PMC article.
-
Helix-7 in Argonaute2 shapes the microRNA seed region for rapid target recognition.EMBO J. 2018 Jan 4;37(1):75-88. doi: 10.15252/embj.201796474. Epub 2017 Sep 22. EMBO J. 2018. PMID: 28939659 Free PMC article.
-
Phosphorylation of Argonaute proteins affects mRNA binding and is essential for microRNA-guided gene silencing in vivo.EMBO J. 2017 Jul 14;36(14):2088-2106. doi: 10.15252/embj.201696386. Epub 2017 Jun 23. EMBO J. 2017. PMID: 28645918 Free PMC article.
-
New insights into the function of mammalian Argonaute2.PLoS Genet. 2020 Nov 12;16(11):e1009058. doi: 10.1371/journal.pgen.1009058. eCollection 2020 Nov. PLoS Genet. 2020. PMID: 33180792 Free PMC article. Review.
-
Why Argonaute is needed to make microRNA target search fast and reliable.Semin Cell Dev Biol. 2017 May;65:20-28. doi: 10.1016/j.semcdb.2016.05.017. Epub 2016 May 26. Semin Cell Dev Biol. 2017. PMID: 27235676 Review.
Cited by
-
Lipid nanovehicles overcome barriers to systemic RNA delivery: Lipid components, fabrication methods, and rational design.Acta Pharm Sin B. 2024 Feb;14(2):579-601. doi: 10.1016/j.apsb.2023.10.012. Epub 2023 Oct 20. Acta Pharm Sin B. 2024. PMID: 38322344 Free PMC article. Review.
-
Ribosome Recycling by ABCE1 Links Lysosomal Function and Iron Homeostasis to 3' UTR-Directed Regulation and Nonsense-Mediated Decay.Cell Rep. 2020 Jul 14;32(2):107895. doi: 10.1016/j.celrep.2020.107895. Cell Rep. 2020. PMID: 32668236 Free PMC article.
-
Robust partitioning of microRNA targets from downstream regulatory changes.Nucleic Acids Res. 2020 Sep 25;48(17):9724-9746. doi: 10.1093/nar/gkaa687. Nucleic Acids Res. 2020. PMID: 32821933 Free PMC article.
-
A specific type of Argonaute phosphorylation regulates binding to microRNAs during C. elegans development.Cell Rep. 2022 Dec 13;41(11):111822. doi: 10.1016/j.celrep.2022.111822. Cell Rep. 2022. PMID: 36516777 Free PMC article.
-
Expression of TNRC6 (GW182) Proteins Is Not Necessary for Gene Silencing by Fully Complementary RNA Duplexes.Nucleic Acid Ther. 2019 Dec;29(6):323-334. doi: 10.1089/nat.2019.0815. Epub 2019 Oct 31. Nucleic Acid Ther. 2019. PMID: 31670606 Free PMC article.
References
-
- Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16:421–433. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

