Individualised multiplexed circulating tumour DNA assays for monitoring of tumour presence in patients after colorectal cancer surgery
- PMID: 28102343
- PMCID: PMC5244357
- DOI: 10.1038/srep40737
Individualised multiplexed circulating tumour DNA assays for monitoring of tumour presence in patients after colorectal cancer surgery
Abstract
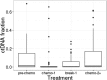
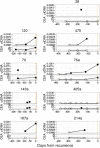
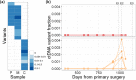
Circulating tumour DNA (ctDNA) has the potential to be a specific biomarker for the monitoring of tumours in patients with colorectal cancer (CRC). Here, our aim was to develop a personalised surveillance strategy to monitor the clinical course of CRC after surgery. We developed patient-specific ctDNA assays based on multiplexed detection of somatic mutations identified from patient primary tumours, and applied them to detect ctDNA in 44 CRC patients, analysing a total of 260 plasma samples. We found that ctDNA detection correlated with clinical events - it is detectable in pre-operative but not post-operative plasma, and also in patients with recurrent CRC. We also detected ctDNA in 11 out of 15 cases at or before clinical or radiological recurrence of CRC, indicating the potential of our assay for early detection of metastasis. We further present data from a patient with multiple primary cancers to demonstrate the specificity of our assays to distinguish between CRC recurrence and a second primary cancer. Our approach can complement current methods for surveillance of CRC by adding an individualised biological component, allowing us not only to point to the presence of residual or recurrent disease, but also attribute it to the original cancer.
Figures
Similar articles
-
Relationship between post-surgery detection of methylated circulating tumor DNA with risk of residual disease and recurrence-free survival.J Cancer Res Clin Oncol. 2018 Sep;144(9):1741-1750. doi: 10.1007/s00432-018-2701-x. Epub 2018 Jul 10. J Cancer Res Clin Oncol. 2018. PMID: 29992492
-
Multiplex detection of ctDNA mutations in plasma of colorectal cancer patients by PCR/SERS assay.Nanotheranostics. 2020 Aug 25;4(4):224-232. doi: 10.7150/ntno.48905. eCollection 2020. Nanotheranostics. 2020. PMID: 32923312 Free PMC article.
-
Frequent post-operative monitoring of colorectal cancer using individualised ctDNA validated by multiregional molecular profiling.Br J Cancer. 2021 Apr;124(9):1556-1565. doi: 10.1038/s41416-021-01266-4. Epub 2021 Mar 3. Br J Cancer. 2021. PMID: 33658639 Free PMC article.
-
[Liquid biopsy in colorectal cancer : An overview of ctDNA analysis in tumour diagnostics].Pathologe. 2019 Dec;40(Suppl 3):244-251. doi: 10.1007/s00292-019-00698-3. Pathologe. 2019. PMID: 31797045 Review. German.
-
Clinical application of circulating tumour DNA in colorectal cancer.Lancet Gastroenterol Hepatol. 2023 Sep;8(9):837-852. doi: 10.1016/S2468-1253(23)00146-2. Epub 2023 Jul 24. Lancet Gastroenterol Hepatol. 2023. PMID: 37499673 Review.
Cited by
-
Relationship between post-surgery detection of methylated circulating tumor DNA with risk of residual disease and recurrence-free survival.J Cancer Res Clin Oncol. 2018 Sep;144(9):1741-1750. doi: 10.1007/s00432-018-2701-x. Epub 2018 Jul 10. J Cancer Res Clin Oncol. 2018. PMID: 29992492
-
LiquidCNA: Tracking subclonal evolution from longitudinal liquid biopsies using somatic copy number alterations.iScience. 2021 Jul 21;24(8):102889. doi: 10.1016/j.isci.2021.102889. eCollection 2021 Aug 20. iScience. 2021. PMID: 34401670 Free PMC article.
-
Advantages and Challenges of Using ctDNA NGS to Assess the Presence of Minimal Residual Disease (MRD) in Solid Tumors.Cancers (Basel). 2021 Nov 14;13(22):5698. doi: 10.3390/cancers13225698. Cancers (Basel). 2021. PMID: 34830853 Free PMC article. Review.
-
Comparison of Somatic Mutation Profiles Between Formalin-Fixed Paraffin Embedded Tissues and Plasma Cell-Free DNA from Ovarian Cancer Patients Before and After Surgery.Biores Open Access. 2020 Mar 13;9(1):73-79. doi: 10.1089/biores.2019.0031. eCollection 2020. Biores Open Access. 2020. PMID: 32219013 Free PMC article.
-
Kinetics of plasma cfDNA predicts clinical response in non-small cell lung cancer patients.Sci Rep. 2021 Apr 7;11(1):7633. doi: 10.1038/s41598-021-85797-z. Sci Rep. 2021. PMID: 33828112 Free PMC article.
References
-
- Labianca R., Nordlinger B., Beretta G. D., Brouquet A. & Cervantes A. Primary colon cancer: ESMO clinical practice guidelines for diagnosis, adjuvant treatment and follow-up. Ann. Oncol. 21, 70–77 (2010). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

