Uncovering the SUMOylation and ubiquitylation crosstalk in human cells using sequential peptide immunopurification
- PMID: 28098164
- PMCID: PMC5253644
- DOI: 10.1038/ncomms14109
Uncovering the SUMOylation and ubiquitylation crosstalk in human cells using sequential peptide immunopurification
Abstract
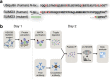
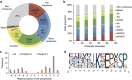
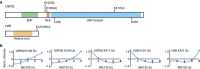
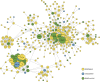
Crosstalk between the SUMO and ubiquitin pathways has recently been reported. However, no approach currently exists to determine the interrelationship between these modifications. Here, we report an optimized immunoaffinity method that permits the study of both protein ubiquitylation and SUMOylation from a single sample. This method enables the unprecedented identification of 10,388 SUMO sites in HEK293 cells. The sequential use of SUMO and ubiquitin remnant immunoaffinity purification facilitates the dynamic profiling of SUMOylated and ubiquitylated proteins in HEK293 cells treated with the proteasome inhibitor MG132. Quantitative proteomic analyses reveals crosstalk between substrates that control protein degradation, and highlights co-regulation of SUMOylation and ubiquitylation levels on deubiquitinase enzymes and the SUMOylation of proteasome subunits. The SUMOylation of the proteasome affects its recruitment to promyelocytic leukemia protein (PML) nuclear bodies, and PML lacking the SUMO interacting motif fails to colocalize with SUMOylated proteasome further demonstrating that this motif is required for PML catabolism.
Figures

Similar articles
-
Inhibiting ubiquitination causes an accumulation of SUMOylated newly synthesized nuclear proteins at PML bodies.J Biol Chem. 2019 Oct 18;294(42):15218-15234. doi: 10.1074/jbc.RA119.009147. Epub 2019 Jul 8. J Biol Chem. 2019. PMID: 31285264 Free PMC article.
-
c-Myc is targeted to the proteasome for degradation in a SUMOylation-dependent manner, regulated by PIAS1, SENP7 and RNF4.Cell Cycle. 2015;14(12):1859-72. doi: 10.1080/15384101.2015.1040965. Cell Cycle. 2015. PMID: 25895136 Free PMC article.
-
Targeted identification of SUMOylation sites in human proteins using affinity enrichment and paralog-specific reporter ions.Mol Cell Proteomics. 2013 Sep;12(9):2536-50. doi: 10.1074/mcp.M112.025569. Epub 2013 Jun 7. Mol Cell Proteomics. 2013. PMID: 23750026 Free PMC article.
-
Interferon, restriction factors and SUMO pathways.Cytokine Growth Factor Rev. 2020 Oct;55:37-47. doi: 10.1016/j.cytogfr.2020.03.001. Epub 2020 May 8. Cytokine Growth Factor Rev. 2020. PMID: 32591223 Review.
-
Gammaherpesviral Tegument Proteins, PML-Nuclear Bodies and the Ubiquitin-Proteasome System.Viruses. 2017 Oct 21;9(10):308. doi: 10.3390/v9100308. Viruses. 2017. PMID: 29065450 Free PMC article. Review.
Cited by
-
SENP2 promotes ESCC proliferation through SETDB1 deSUMOylation and enhanced fatty acid metabolism.Heliyon. 2024 Jul 2;10(13):e34010. doi: 10.1016/j.heliyon.2024.e34010. eCollection 2024 Jul 15. Heliyon. 2024. PMID: 39071660 Free PMC article.
-
SUMO protease SENP1 deSUMOylates and stabilizes c-Myc.Proc Natl Acad Sci U S A. 2018 Oct 23;115(43):10983-10988. doi: 10.1073/pnas.1802932115. Epub 2018 Oct 10. Proc Natl Acad Sci U S A. 2018. PMID: 30305424 Free PMC article.
-
SUMO-Mediated Regulation of Nuclear Functions and Signaling Processes.Mol Cell. 2018 Aug 2;71(3):409-418. doi: 10.1016/j.molcel.2018.07.027. Mol Cell. 2018. PMID: 30075142 Free PMC article. Review.
-
Targeting the MYC Ubiquitination-Proteasome Degradation Pathway for Cancer Therapy.Front Oncol. 2021 Jun 11;11:679445. doi: 10.3389/fonc.2021.679445. eCollection 2021. Front Oncol. 2021. PMID: 34178666 Free PMC article. Review.
-
Src SUMOylation Inhibits Tumor Growth Via Decreasing FAK Y925 Phosphorylation.Neoplasia. 2017 Dec;19(12):961-971. doi: 10.1016/j.neo.2017.09.001. Epub 2017 Oct 22. Neoplasia. 2017. PMID: 29069627 Free PMC article.
References
-
- Geoffroy M.-C. & Hay R. T. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat. Rev. Mol. Cell. Biol. 10, 564–568 (2009). - PubMed
-
- Johnson E. S. Protein modification by SUMO. Annu. Rev. Biochem. 73, 355–382 (2004). - PubMed
-
- Melchior F. SUMO--nonclassical ubiquitin. Annu. Rev. Cell. Dev. Biol. 16, 591–626 (2000). - PubMed
-
- Saitoh H. & Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275, 6252–6258 (2000). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

