MAHMI database: a comprehensive MetaHit-based resource for the study of the mechanism of action of the human microbiota
- PMID: 28077565
- PMCID: PMC5225402
- DOI: 10.1093/database/baw157
MAHMI database: a comprehensive MetaHit-based resource for the study of the mechanism of action of the human microbiota
Abstract
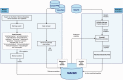
The Mechanism of Action of the Human Microbiome (MAHMI) database is a unique resource that provides comprehensive information about the sequence of potential immunomodulatory and antiproliferative peptides encrypted in the proteins produced by the human gut microbiota. Currently, MAHMI database contains over 300 hundred million peptide entries, with detailed information about peptide sequence, sources and potential bioactivity. The reference peptide data section is curated manually by domain experts. The in silico peptide data section is populated automatically through the systematic processing of publicly available exoproteomes of the human microbiome. Bioactivity prediction is based on the global alignment of the automatically processed peptides with experimentally validated immunomodulatory and antiproliferative peptides, in the reference section. MAHMI provides researchers with a comparative tool for inspecting the potential immunomodulatory or antiproliferative bioactivity of new amino acidic sequences and identifying promising peptides to be further investigated. Moreover, researchers are welcome to submit new experimental evidence on peptide bioactivity, namely, empiric and structural data, as a proactive, expert means to keep the database updated and improve the implemented bioactivity prediction method. Bioactive peptides identified by MAHMI have a huge biotechnological potential, including the manipulation of aberrant immune responses and the design of new functional ingredients/foods based on the genetic sequences of the human microbiome. Hopefully, the resources provided by MAHMI will be useful to those researching gastrointestinal disorders of autoimmune and inflammatory nature, such as Inflammatory Bowel Diseases. MAHMI database is routinely updated and is available free of charge. Database URL: http://mahmi.org/.
© The Author(s) 2017. Published by Oxford University Press.
Figures

Similar articles
-
In Silico Screening of the Human Gut Metaproteome Identifies Th17-Promoting Peptides Encrypted in Proteins of Commensal Bacteria.Front Microbiol. 2017 Sep 8;8:1726. doi: 10.3389/fmicb.2017.01726. eCollection 2017. Front Microbiol. 2017. PMID: 28943872 Free PMC article.
-
In silico prediction reveals the existence of potential bioactive neuropeptides produced by the human gut microbiota.Food Res Int. 2019 May;119:221-226. doi: 10.1016/j.foodres.2019.01.069. Epub 2019 Jan 30. Food Res Int. 2019. PMID: 30884651
-
SpirPep: an in silico digestion-based platform to assist bioactive peptides discovery from a genome-wide database.BMC Bioinformatics. 2018 Apr 20;19(1):149. doi: 10.1186/s12859-018-2143-0. BMC Bioinformatics. 2018. PMID: 29678128 Free PMC article.
-
Integration of microbiome and epigenome to decipher the pathogenesis of autoimmune diseases.J Autoimmun. 2017 Sep;83:31-42. doi: 10.1016/j.jaut.2017.03.009. Epub 2017 Mar 23. J Autoimmun. 2017. PMID: 28342734 Review.
-
The human microbiome in rheumatic autoimmune diseases: A comprehensive review.Clin Immunol. 2016 Sep;170:70-9. doi: 10.1016/j.clim.2016.07.026. Epub 2016 Aug 2. Clin Immunol. 2016. PMID: 27493014 Review.
Cited by
-
A Metabolomics Approach Reveals Immunomodulatory Effects of Proteinaceous Molecules Derived From Gut Bacteria Over Human Peripheral Blood Mononuclear Cells.Front Microbiol. 2018 Nov 13;9:2701. doi: 10.3389/fmicb.2018.02701. eCollection 2018. Front Microbiol. 2018. PMID: 30524384 Free PMC article.
-
Immunomodulatory Effect of Gut Microbiota-Derived Bioactive Peptides on Human Immune System from Healthy Controls and Patients with Inflammatory Bowel Disease.Nutrients. 2019 Oct 31;11(11):2605. doi: 10.3390/nu11112605. Nutrients. 2019. PMID: 31683517 Free PMC article.
-
Worldwide trends in scientific publications on association of gut microbiota with obesity.Iran J Basic Med Sci. 2019 Jan;22(1):65-71. doi: 10.22038/ijbms.2018.30203.7281. Iran J Basic Med Sci. 2019. PMID: 30944710 Free PMC article. Review.
-
In Silico Screening of the Human Gut Metaproteome Identifies Th17-Promoting Peptides Encrypted in Proteins of Commensal Bacteria.Front Microbiol. 2017 Sep 8;8:1726. doi: 10.3389/fmicb.2017.01726. eCollection 2017. Front Microbiol. 2017. PMID: 28943872 Free PMC article.
-
Microbiota-Derived Metabolites, Indole-3-aldehyde and Indole-3-acetic Acid, Differentially Modulate Innate Cytokines and Stromal Remodeling Processes Associated with Autoimmune Arthritis.Int J Mol Sci. 2021 Feb 18;22(4):2017. doi: 10.3390/ijms22042017. Int J Mol Sci. 2021. PMID: 33670600 Free PMC article.
References
-
- Hooper L.V., Gordon J.I. (2001) Commensal host-bacterial relationships in the gut. Science, 292, 1115–1118. - PubMed
-
- Sommer F., Bäckhed F. (2013) The gut microbiota—masters of host development and physiology. Nat. Rev. Microbiol., 11, 227–238. - PubMed
-
- Sánchez B., Urdaci M.C., Margolles A. (2010) Extracellular proteins secreted by probiotic bacteria as mediators of effects that promote mucosa-bacteria interactions. Microbiology, 156, 3232–3242. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

