Accurate De Novo Prediction of Protein Contact Map by Ultra-Deep Learning Model
- PMID: 28056090
- PMCID: PMC5249242
- DOI: 10.1371/journal.pcbi.1005324
Accurate De Novo Prediction of Protein Contact Map by Ultra-Deep Learning Model
Abstract
Motivation: Protein contacts contain key information for the understanding of protein structure and function and thus, contact prediction from sequence is an important problem. Recently exciting progress has been made on this problem, but the predicted contacts for proteins without many sequence homologs is still of low quality and not very useful for de novo structure prediction.
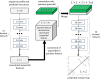
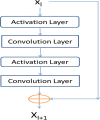
Method: This paper presents a new deep learning method that predicts contacts by integrating both evolutionary coupling (EC) and sequence conservation information through an ultra-deep neural network formed by two deep residual neural networks. The first residual network conducts a series of 1-dimensional convolutional transformation of sequential features; the second residual network conducts a series of 2-dimensional convolutional transformation of pairwise information including output of the first residual network, EC information and pairwise potential. By using very deep residual networks, we can accurately model contact occurrence patterns and complex sequence-structure relationship and thus, obtain higher-quality contact prediction regardless of how many sequence homologs are available for proteins in question.
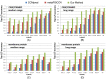
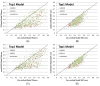
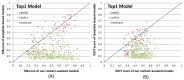
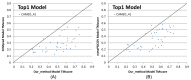
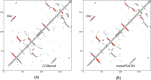
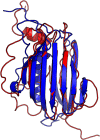
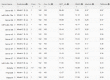
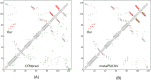
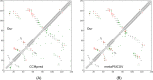
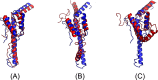
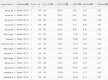
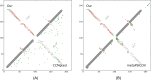
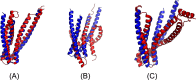
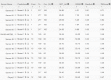
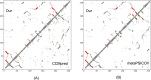
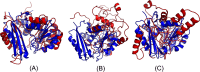
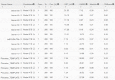
Results: Our method greatly outperforms existing methods and leads to much more accurate contact-assisted folding. Tested on 105 CASP11 targets, 76 past CAMEO hard targets, and 398 membrane proteins, the average top L long-range prediction accuracy obtained by our method, one representative EC method CCMpred and the CASP11 winner MetaPSICOV is 0.47, 0.21 and 0.30, respectively; the average top L/10 long-range accuracy of our method, CCMpred and MetaPSICOV is 0.77, 0.47 and 0.59, respectively. Ab initio folding using our predicted contacts as restraints but without any force fields can yield correct folds (i.e., TMscore>0.6) for 203 of the 579 test proteins, while that using MetaPSICOV- and CCMpred-predicted contacts can do so for only 79 and 62 of them, respectively. Our contact-assisted models also have much better quality than template-based models especially for membrane proteins. The 3D models built from our contact prediction have TMscore>0.5 for 208 of the 398 membrane proteins, while those from homology modeling have TMscore>0.5 for only 10 of them. Further, even if trained mostly by soluble proteins, our deep learning method works very well on membrane proteins. In the recent blind CAMEO benchmark, our fully-automated web server implementing this method successfully folded 6 targets with a new fold and only 0.3L-2.3L effective sequence homologs, including one β protein of 182 residues, one α+β protein of 125 residues, one α protein of 140 residues, one α protein of 217 residues, one α/β of 260 residues and one α protein of 462 residues. Our method also achieved the highest F1 score on free-modeling targets in the latest CASP (Critical Assessment of Structure Prediction), although it was not fully implemented back then.
Availability: http://raptorx.uchicago.edu/ContactMap/.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

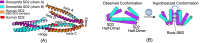
Similar articles
-
Analysis of distance-based protein structure prediction by deep learning in CASP13.Proteins. 2019 Dec;87(12):1069-1081. doi: 10.1002/prot.25810. Epub 2019 Sep 13. Proteins. 2019. PMID: 31471916
-
MetaPSICOV: combining coevolution methods for accurate prediction of contacts and long range hydrogen bonding in proteins.Bioinformatics. 2015 Apr 1;31(7):999-1006. doi: 10.1093/bioinformatics/btu791. Epub 2014 Nov 26. Bioinformatics. 2015. PMID: 25431331 Free PMC article.
-
DNCON2: improved protein contact prediction using two-level deep convolutional neural networks.Bioinformatics. 2018 May 1;34(9):1466-1472. doi: 10.1093/bioinformatics/btx781. Bioinformatics. 2018. PMID: 29228185 Free PMC article.
-
A decade of CASP: progress, bottlenecks and prognosis in protein structure prediction.Curr Opin Struct Biol. 2005 Jun;15(3):285-9. doi: 10.1016/j.sbi.2005.05.011. Curr Opin Struct Biol. 2005. PMID: 15939584 Review.
-
Machine learning in protein structure prediction.Curr Opin Chem Biol. 2021 Dec;65:1-8. doi: 10.1016/j.cbpa.2021.04.005. Epub 2021 May 18. Curr Opin Chem Biol. 2021. PMID: 34015749 Review.
Cited by
-
DeepSRE: Identification of sterol responsive elements and nuclear transcription factors Y proximity in human DNA by Convolutional Neural Network analysis.PLoS One. 2021 Mar 4;16(3):e0247402. doi: 10.1371/journal.pone.0247402. eCollection 2021. PLoS One. 2021. PMID: 33661949 Free PMC article.
-
Deep Learning in Proteomics.Proteomics. 2020 Nov;20(21-22):e1900335. doi: 10.1002/pmic.201900335. Epub 2020 Oct 30. Proteomics. 2020. PMID: 32939979 Free PMC article. Review.
-
Addressing epistasis in the design of protein function.Proc Natl Acad Sci U S A. 2024 Aug 20;121(34):e2314999121. doi: 10.1073/pnas.2314999121. Epub 2024 Aug 12. Proc Natl Acad Sci U S A. 2024. PMID: 39133844 Free PMC article.
-
Deep learning for protein structure prediction and design-progress and applications.Mol Syst Biol. 2024 Mar;20(3):162-169. doi: 10.1038/s44320-024-00016-x. Epub 2024 Jan 30. Mol Syst Biol. 2024. PMID: 38291232 Free PMC article. Review.
-
Improved inter-residue contact prediction via a hybrid generative model and dynamic loss function.Comput Struct Biotechnol J. 2022 Nov 12;20:6138-6148. doi: 10.1016/j.csbj.2022.11.020. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36420166 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

