How Does Your Protein Fold? Elucidating the Apomyoglobin Folding Pathway
- PMID: 28032989
- PMCID: PMC5241236
- DOI: 10.1021/acs.accounts.6b00511
How Does Your Protein Fold? Elucidating the Apomyoglobin Folding Pathway
Abstract
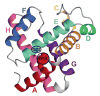
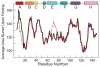
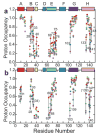
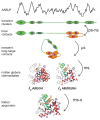
Although each type of protein fold and in some cases individual proteins within a fold classification can have very different mechanisms of folding, the underlying biophysical and biochemical principles that operate to cause a linear polypeptide chain to fold into a globular structure must be the same. In an aqueous solution, the protein takes up the thermodynamically most stable structure, but the pathway along which the polypeptide proceeds in order to reach that structure is a function of the amino acid sequence, which must be the final determining factor, not only in shaping the final folded structure, but in dictating the folding pathway. A number of groups have focused on a single protein or group of proteins, to determine in detail the factors that influence the rate and mechanism of folding in a defined system, with the hope that hypothesis-driven experiments can elucidate the underlying principles governing the folding process. Our research group has focused on the folding of the globin family of proteins, and in particular on the monomeric protein apomyoglobin. Apomyoglobin (apoMb) folds relatively slowly (∼2 s) via an ensemble of obligatory intermediates that form rapidly after the initiation of folding. The folding pathway can be dissected using rapid-mixing techniques, which can probe processes in the millisecond time range. Stopped-flow measurements detected by circular dichroism (CD) or fluorescence spectroscopy give information on the rates of folding events. Quench-flow experiments utilize the differential rates of hydrogen-deuterium exchange of amide protons protected in parts of the structure that are folded early; protection of amides can be detected by mass spectrometry or proton nuclear magnetic resonance spectroscopy (NMR). In addition, apoMb forms an intermediate at equilibrium at pH ∼ 4, which is sufficiently stable for it to be structurally characterized by solution methods such as CD, fluorescence and NMR spectroscopies, and the conformational ensembles formed in the presence of denaturing agents and low pH can be characterized as models for the unfolded states of the protein. Newer NMR techniques such as measurement of residual dipolar couplings in the various partly folded states, and relaxation dispersion measurements to probe invisible states present at low concentrations, have contributed to providing a detailed picture of the apomyoglobin folding pathway. The research summarized in this Account was aimed at characterizing and comparing the equilibrium and kinetic intermediates both structurally and dynamically, as well as delineating the complete folding pathway at a residue-specific level, in order to answer the question: "What is it about the amino acid sequence that causes each molecule in the unfolded protein ensemble to start folding, and, once started, to proceed towards the formation of the correctly folded three-dimensional structure?"
Figures
Similar articles
-
Effect of H helix destabilizing mutations on the kinetic and equilibrium folding of apomyoglobin.J Mol Biol. 1999 Jan 8;285(1):269-82. doi: 10.1006/jmbi.1998.2273. J Mol Biol. 1999. PMID: 9878405
-
The kinetic and equilibrium molten globule intermediates of apoleghemoglobin differ in structure.J Mol Biol. 2008 May 2;378(3):715-25. doi: 10.1016/j.jmb.2008.03.025. Epub 2008 Mar 19. J Mol Biol. 2008. PMID: 18384808 Free PMC article.
-
Structural characterization of unfolded states of apomyoglobin using residual dipolar couplings.J Mol Biol. 2004 Jul 23;340(5):1131-42. doi: 10.1016/j.jmb.2004.05.022. J Mol Biol. 2004. PMID: 15236972
-
Folding of apomyoglobin: Analysis of transient intermediate structure during refolding using quick hydrogen deuterium exchange and NMR.Proc Jpn Acad Ser B Phys Biol Sci. 2017;93(1):10-27. doi: 10.2183/pjab.93.002. Proc Jpn Acad Ser B Phys Biol Sci. 2017. PMID: 28077807 Free PMC article. Review.
-
The folding process of apomyoglobin.Protein Pept Lett. 2005 Apr;12(3):229-34. doi: 10.2174/0929866053587174. Protein Pept Lett. 2005. PMID: 15777270 Review.
Cited by
-
Mapping Protein-Protein Interactions at Birth: Single-Particle Cryo-EM Analysis of a Ribosome-Nascent Globin Complex.ACS Cent Sci. 2024 Feb 1;10(2):385-401. doi: 10.1021/acscentsci.3c00777. eCollection 2024 Feb 28. ACS Cent Sci. 2024. PMID: 38435509 Free PMC article.
-
Water stabilizes an alternate turn conformation in horse heart myoglobin.Sci Rep. 2023 Apr 13;13(1):6094. doi: 10.1038/s41598-023-32821-z. Sci Rep. 2023. PMID: 37055458 Free PMC article.
-
Chemical Exchange.Methods Enzymol. 2019;615:177-236. doi: 10.1016/bs.mie.2018.09.028. Epub 2018 Dec 4. Methods Enzymol. 2019. PMID: 30638530 Free PMC article.
-
Crowding-Induced Elongated Conformation of Urea-Unfolded Apoazurin: Investigating the Role of Crowder Shape in Silico.J Phys Chem B. 2019 May 2;123(17):3607-3617. doi: 10.1021/acs.jpcb.9b00782. Epub 2019 Apr 23. J Phys Chem B. 2019. PMID: 30963769 Free PMC article.
-
Retrospective study for the universal applicability of the residue-based linear free energy relationship in the two-state exchange of protein molecules.Sci Rep. 2022 Oct 7;12(1):16843. doi: 10.1038/s41598-022-21226-z. Sci Rep. 2022. PMID: 36207470 Free PMC article.
References
-
- Jamin M, Yeh SR, Rousseau DL, Baldwin RL. Submillisecond unfolding kinetics of apomyoglobin and its pH 4 intermediate. J Mol Biol. 1999;292:731–740. - PubMed
-
- Gulotta M, Gilmanshin R, Buscher TC, Callender RH, Dyer RB. Core formation in apomyoglobin: probing the upper reaches of the folding energy landscape. Biochemistry. 2001;40:5137–5143. - PubMed
-
- Griko YV, Privalov PL, Venyaminov SY, Kutyshenko VP. Thermodynamic study of the apomyoglobin structure. J Mol Biol. 1988;202:127–138. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

