MicroRNA-202 maintains spermatogonial stem cells by inhibiting cell cycle regulators and RNA binding proteins
- PMID: 27998933
- PMCID: PMC5397178
- DOI: 10.1093/nar/gkw1287
MicroRNA-202 maintains spermatogonial stem cells by inhibiting cell cycle regulators and RNA binding proteins
Abstract
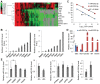
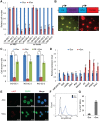
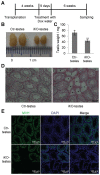
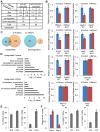
miRNAs play important roles during mammalian spermatogenesis. However, the function of most miRNAs in spermatogenesis and the underlying mechanisms remain unknown. Here, we report that miR-202 is highly expressed in mouse spermatogonial stem cells (SSCs), and is oppositely regulated by Glial cell-Derived Neurotrophic Factor (GDNF) and retinoic acid (RA), two key factors for SSC self-renewal and differentiation. We used inducible CRISPR-Cas9 to knockout miR-202 in cultured SSCs, and found that the knockout SSCs initiated premature differentiation accompanied by reduced stem cell activity and increased mitosis and apoptosis. Target genes were identified with iTRAQ-based proteomic analysis and RNA sequencing, and are enriched with cell cycle regulators and RNA-binding proteins. Rbfox2 and Cpeb1 were found to be direct targets of miR-202 and Rbfox2 but not Cpeb1, is essential for the differentiation of SSCs into meiotic cells. Accordingly, an SSC fate-regulatory network composed of signaling molecules of GDNF and RA, miR-202 and diverse downstream effectors has been identified.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
A Novel Regulatory Axis, CHD1L-MicroRNA 486-Matrix Metalloproteinase 2, Controls Spermatogonial Stem Cell Properties.Mol Cell Biol. 2019 Feb 4;39(4):e00357-18. doi: 10.1128/MCB.00357-18. Print 2019 Feb 15. Mol Cell Biol. 2019. PMID: 30455250 Free PMC article.
-
The microRNA miR-202 prevents precocious spermatogonial differentiation and meiotic initiation during mouse spermatogenesis.Development. 2021 Dec 15;148(24):dev199799. doi: 10.1242/dev.199799. Epub 2021 Dec 16. Development. 2021. PMID: 34913465
-
MicroRNA-30a-5p promotes differentiation in neonatal mouse spermatogonial stem cells (SSCs).Reprod Biol Endocrinol. 2021 Jun 9;19(1):85. doi: 10.1186/s12958-021-00758-5. Reprod Biol Endocrinol. 2021. PMID: 34108007 Free PMC article.
-
Noncoding RNAs: Potential players in the self-renewal of mammalian spermatogonial stem cells.Mol Reprod Dev. 2018 Aug;85(8-9):720-728. doi: 10.1002/mrd.23041. Epub 2018 Aug 16. Mol Reprod Dev. 2018. PMID: 29969526 Review.
-
The involvement of bioactive factors in the self-renewal and stemness maintenance of spermatogonial stem cells.Mol Cell Biochem. 2021 Apr;476(4):1813-1823. doi: 10.1007/s11010-020-04028-7. Epub 2021 Jan 18. Mol Cell Biochem. 2021. PMID: 33459979 Review.
Cited by
-
Epididymal epithelium propels early sexual transmission of Zika virus in the absence of interferon signaling.Nat Commun. 2021 Apr 29;12(1):2469. doi: 10.1038/s41467-021-22729-5. Nat Commun. 2021. PMID: 33927207 Free PMC article.
-
Loss of circSRY reduces γH2AX level in germ cells and impairs mouse spermatogenesis.Life Sci Alliance. 2022 Nov 22;6(2):e202201617. doi: 10.26508/lsa.202201617. Print 2023 Feb. Life Sci Alliance. 2022. PMID: 36414375 Free PMC article.
-
In vivo functions of miRNAs in mammalian spermatogenesis.Front Cell Dev Biol. 2023 May 5;11:1154938. doi: 10.3389/fcell.2023.1154938. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37215089 Free PMC article. Review.
-
Splicing regulation in brain and testis: common themes for highly specialized organs.Cell Cycle. 2021 Mar-Mar;20(5-6):480-489. doi: 10.1080/15384101.2021.1889187. Epub 2021 Feb 26. Cell Cycle. 2021. PMID: 33632061 Free PMC article. Review.
-
The Role of microRNA in Spermatogenesis: Is There a Place for Fertility Preservation Innovation?Int J Mol Sci. 2023 Dec 29;25(1):460. doi: 10.3390/ijms25010460. Int J Mol Sci. 2023. PMID: 38203631 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

