Receptor Activation of HIV-1 Env Leads to Asymmetric Exposure of the gp41 Trimer
- PMID: 27992602
- PMCID: PMC5222517
- DOI: 10.1371/journal.ppat.1006098
Receptor Activation of HIV-1 Env Leads to Asymmetric Exposure of the gp41 Trimer
Abstract
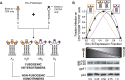
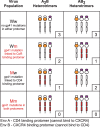
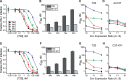
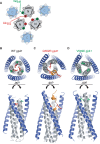
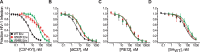
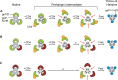
Structural rearrangements of HIV-1 glycoprotein Env promote viral entry through membrane fusion. Env is a symmetric homotrimer with each protomer composed of surface subunit gp120 and transmembrane subunit gp41. Cellular CD4- and chemokine receptor-binding to gp120 coordinate conformational changes in gp41, first to an extended prehairpin intermediate (PHI) and, ultimately, into a fusogenic trimer-of-hairpins (TOH). HIV-1 fusion inhibitors target gp41 in the PHI and block TOH formation. To characterize structural transformations into and through the PHI, we employed asymmetric Env trimers containing both high and low affinity binding sites for individual fusion inhibitors. Asymmetry was achieved using engineered Env heterotrimers composed of protomers deficient in either CD4- or chemokine receptor-binding. Linking receptor engagement to inhibitor affinity allowed us to assess conformational changes of individual Env protomers in the context of a functioning trimer. We found that the transition into the PHI could occur symmetrically or asymmetrically depending on the stoichiometry of CD4 binding. Sequential engagement of multiple CD4s promoted progressive exposure of individual fusion inhibitor binding sites in a CD4-dependent fashion. By contrast, engagement of only a single CD4 molecule led to a delayed, but symmetric, exposure of the gp41 trimer. This complex coupling between Env-CD4 interaction and gp41 exposure explained the multiphasic fusion-inhibitor titration observed for a mutant Env homotrimer with a naturally asymmetric gp41. Our results suggest that the spatial and temporal exposure of gp41 can proceed in a nonconcerted, asymmetric manner depending on the number of CD4s that engage the Env trimer. The findings have important implications for the mechanism of viral membrane fusion and the development of vaccine candidates designed to elicit neutralizing antibodies targeting gp41 in the PHI.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
HIV-1 gp41 Residues Modulate CD4-Induced Conformational Changes in the Envelope Glycoprotein and Evolution of a Relaxed Conformation of gp120.J Virol. 2018 Jul 31;92(16):e00583-18. doi: 10.1128/JVI.00583-18. Print 2018 Aug 15. J Virol. 2018. PMID: 29875245 Free PMC article.
-
Residues in the gp41 Ectodomain Regulate HIV-1 Envelope Glycoprotein Conformational Transitions Induced by gp120-Directed Inhibitors.J Virol. 2017 Feb 14;91(5):e02219-16. doi: 10.1128/JVI.02219-16. Print 2017 Mar 1. J Virol. 2017. PMID: 28003492 Free PMC article.
-
Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop.Proc Natl Acad Sci U S A. 2016 Nov 15;113(46):E7151-E7158. doi: 10.1073/pnas.1615939113. Epub 2016 Oct 31. Proc Natl Acad Sci U S A. 2016. PMID: 27799557 Free PMC article.
-
Quaternary Interaction of the HIV-1 Envelope Trimer with CD4 and Neutralizing Antibodies.Viruses. 2021 Jul 20;13(7):1405. doi: 10.3390/v13071405. Viruses. 2021. PMID: 34372611 Free PMC article. Review.
-
CD4 activation of HIV fusion.Int J Cell Cloning. 1992 Nov;10(6):323-32. doi: 10.1002/stem.5530100603. Int J Cell Cloning. 1992. PMID: 1281202 Review.
Cited by
-
Asymmetric conformations of cleaved HIV-1 envelope glycoprotein trimers in styrene-maleic acid lipid nanoparticles.Commun Biol. 2023 May 18;6(1):535. doi: 10.1038/s42003-023-04916-w. Commun Biol. 2023. PMID: 37202420 Free PMC article.
-
Mutations That Increase the Stability of the Postfusion gp41 Conformation of the HIV-1 Envelope Glycoprotein Are Selected by both an X4 and R5 HIV-1 Virus To Escape Fusion Inhibitors Corresponding to Heptad Repeat 1 of gp41, but the gp120 Adaptive Mutations Differ between the Two Viruses.J Virol. 2019 May 15;93(11):e00142-19. doi: 10.1128/JVI.00142-19. Print 2019 Jun 1. J Virol. 2019. PMID: 30894471 Free PMC article.
-
Global Increases in Human Immunodeficiency Virus Neutralization Sensitivity Due to Alterations in the Membrane-Proximal External Region of the Envelope Glycoprotein Can Be Minimized by Distant State 1-Stabilizing Changes.J Virol. 2022 Apr 13;96(7):e0187821. doi: 10.1128/jvi.01878-21. Epub 2022 Mar 15. J Virol. 2022. PMID: 35289647 Free PMC article.
-
Complex interplay of kinetic factors governs the synergistic properties of HIV-1 entry inhibitors.J Biol Chem. 2017 Oct 6;292(40):16498-16510. doi: 10.1074/jbc.M117.791731. Epub 2017 Jul 10. J Biol Chem. 2017. PMID: 28696261 Free PMC article.
-
Characterization of the Human Immunodeficiency Virus (HIV-1) Envelope Glycoprotein Conformational States on Infectious Virus Particles.J Virol. 2023 Mar 30;97(3):e0185722. doi: 10.1128/jvi.01857-22. Epub 2023 Feb 23. J Virol. 2023. PMID: 36815832 Free PMC article.
References
-
- White TA, Bartesaghi A, Borgnia MJ, Meyerson JR, de la Cruz MJ, Bess JW, et al. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog. 2010;6(12):e1001249 10.1371/journal.ppat.1001249 - DOI - PMC - PubMed
-
- White TA, Bartesaghi A, Borgnia MJ, de la Cruz MJ, Nandwani R, Hoxie JA, et al. Three-dimensional structures of soluble CD4-bound states of trimeric simian immunodeficiency virus envelope glycoproteins determined by using cryo-electron tomography. J Virol. 2011. December;85(23):12114–23. 10.1128/JVI.05297-11 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

