Mutant KRAS Enhances Tumor Cell Fitness by Upregulating Stress Granules
- PMID: 27984728
- PMCID: PMC5441683
- DOI: 10.1016/j.cell.2016.11.035
Mutant KRAS Enhances Tumor Cell Fitness by Upregulating Stress Granules
Abstract
There is growing evidence that stress-coping mechanisms represent tumor cell vulnerabilities that may function as therapeutically beneficial targets. Recent work has delineated an integrated stress adaptation mechanism that is characterized by the formation of cytoplasmic mRNA and protein foci, termed stress granules (SGs). Here, we demonstrate that SGs are markedly elevated in mutant KRAS cells following exposure to stress-inducing stimuli. The upregulation of SGs by mutant KRAS is dependent on the production of the signaling lipid molecule 15-deoxy-delta 12,14 prostaglandin J2 (15-d-PGJ2) and confers cytoprotection against stress stimuli and chemotherapeutic agents. The secretion of 15-d-PGJ2 by mutant KRAS cells is sufficient to enhance SG formation and stress resistance in cancer cells that are wild-type for KRAS. Our findings identify a mutant KRAS-dependent cell non-autonomous mechanism that may afford the establishment of a stress-resistant niche that encompasses different tumor subclones. These results should inform the design of strategies to eradicate tumor cell communities.
Keywords: KRAS; cancer; prostaglandins; stress granules.
Copyright © 2016 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

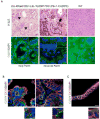
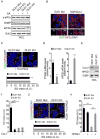

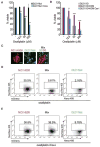
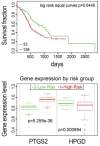
Comment in
-
Mutant RAS Calms Stressed-Out Cancer Cells.Dev Cell. 2017 Jan 23;40(2):120-122. doi: 10.1016/j.devcel.2017.01.005. Dev Cell. 2017. PMID: 28118599 Free PMC article.
Similar articles
-
Targeting eIF4A-Dependent Translation of KRAS Signaling Molecules.Cancer Res. 2021 Apr 15;81(8):2002-2014. doi: 10.1158/0008-5472.CAN-20-2929. Epub 2021 Feb 25. Cancer Res. 2021. PMID: 33632898 Free PMC article.
-
Stress granules are not present in Kras mutant cancers and do not control tumor growth.EMBO Rep. 2024 Nov;25(11):4693-4707. doi: 10.1038/s44319-024-00284-6. Epub 2024 Oct 10. EMBO Rep. 2024. PMID: 39390257 Free PMC article.
-
Evaluating Stress Granules in Pancreatic Cancer In Vitro and In Vivo.Methods Mol Biol. 2019;1882:183-195. doi: 10.1007/978-1-4939-8879-2_17. Methods Mol Biol. 2019. PMID: 30378055
-
Critical role of oncogenic KRAS in pancreatic cancer (Review).Mol Med Rep. 2016 Jun;13(6):4943-9. doi: 10.3892/mmr.2016.5196. Epub 2016 Apr 27. Mol Med Rep. 2016. PMID: 27121414 Review.
-
Synthetic Lethal Vulnerabilities in KRAS-Mutant Cancers.Cold Spring Harb Perspect Med. 2018 Aug 1;8(8):a031518. doi: 10.1101/cshperspect.a031518. Cold Spring Harb Perspect Med. 2018. PMID: 29101114 Free PMC article. Review.
Cited by
-
Stress granules safeguard against MAPK signaling hyperactivation by sequestering PKC/Pck2: new findings and perspectives.Curr Genet. 2021 Dec;67(6):857-863. doi: 10.1007/s00294-021-01192-1. Epub 2021 Jun 7. Curr Genet. 2021. PMID: 34100129 Review.
-
Revisiting the Concept of Stress in the Prognosis of Solid Tumors: A Role for Stress Granules Proteins?Cancers (Basel). 2020 Sep 1;12(9):2470. doi: 10.3390/cancers12092470. Cancers (Basel). 2020. PMID: 32882814 Free PMC article. Review.
-
Stress granules-membraneless organelles as therapeutic targets in pancreatic cancer.EMBO Mol Med. 2024 Mar;16(3):429-431. doi: 10.1038/s44321-024-00040-2. Epub 2024 Feb 27. EMBO Mol Med. 2024. PMID: 38413839 Free PMC article.
-
RAS-mediated tumor stress adaptation and the targeting opportunities it presents.Dis Model Mech. 2022 Feb 1;15(2):dmm049280. doi: 10.1242/dmm.049280. Epub 2022 Feb 11. Dis Model Mech. 2022. PMID: 35147163 Free PMC article.
-
Cellular Stress: Modulator of Regulated Cell Death.Biology (Basel). 2023 Aug 25;12(9):1172. doi: 10.3390/biology12091172. Biology (Basel). 2023. PMID: 37759572 Free PMC article. Review.
References
-
- Abramoff MD, Magalhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics international. 2004;11:36–42.
-
- Aguirre-Gamboa R, Gomez-Rueda H, Martı´nez-Ledesma E, Martı´nez-Torteya A, Chacolla-Huaringa R, Rodriguez-Barrentos A, Tamez-Pen˜ AJG, Trevin˜ OV. SuryExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PloS One. 2013;8:e74250. - PMC - PubMed
-
- Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nature cell biology. 2008;10:1324–1332. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

