Viral DNA Sensors IFI16 and Cyclic GMP-AMP Synthase Possess Distinct Functions in Regulating Viral Gene Expression, Immune Defenses, and Apoptotic Responses during Herpesvirus Infection
- PMID: 27935834
- PMCID: PMC5111403
- DOI: 10.1128/mBio.01553-16
Viral DNA Sensors IFI16 and Cyclic GMP-AMP Synthase Possess Distinct Functions in Regulating Viral Gene Expression, Immune Defenses, and Apoptotic Responses during Herpesvirus Infection
Abstract
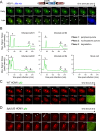
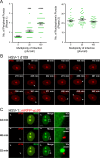
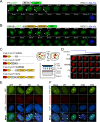
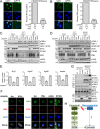
The human interferon-inducible protein IFI16 is an important antiviral factor that binds nuclear viral DNA and promotes antiviral responses. Here, we define IFI16 dynamics in space and time and its distinct functions from the DNA sensor cyclic dinucleotide GMP-AMP synthase (cGAS). Live-cell imaging reveals a multiphasic IFI16 redistribution, first to viral entry sites at the nuclear periphery and then to nucleoplasmic puncta upon herpes simplex virus 1 (HSV-1) and human cytomegalovirus (HCMV) infections. Optogenetics and live-cell microscopy establish the IFI16 pyrin domain as required for nuclear periphery localization and oligomerization. Furthermore, using proteomics, we define the signature protein interactions of the IFI16 pyrin and HIN200 domains and demonstrate the necessity of pyrin for IFI16 interactions with antiviral proteins PML and cGAS. We probe signaling pathways engaged by IFI16, cGAS, and PML using clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9-mediated knockouts in primary fibroblasts. While IFI16 induces cytokines, only cGAS activates STING/TBK-1/IRF3 and apoptotic responses upon HSV-1 and HCMV infections. cGAS-dependent apoptosis upon DNA stimulation requires both the enzymatic production of cyclic dinucleotides and STING. We show that IFI16, not cGAS or PML, represses HSV-1 gene expression, reducing virus titers. This indicates that regulation of viral gene expression may function as a greater barrier to viral replication than the induction of antiviral cytokines. Altogether, our findings establish coordinated and distinct antiviral functions for IFI16 and cGAS against herpesviruses.
Importance: How mammalian cells detect and respond to DNA viruses that replicate in the nucleus is poorly understood. Here, we decipher the distinct functions of two viral DNA sensors, IFI16 and cGAS, during active immune signaling upon infection with two herpesviruses, herpes simplex virus 1 (HSV-1) and human cytomegalovirus (HCMV). We show that IFI16 rapidly oligomerizes at incoming herpesvirus genomes at the nuclear periphery to transcriptionally repress viral gene expression and limit viral replicative capacity. We further demonstrate that IFI16 does not initiate upstream activation of the canonical STING/TBK-1/IRF3 signaling pathway but is required for downstream antiviral cytokine expression. In contrast, we find that, upon DNA sensing during herpesvirus infection, cGAS triggers apoptosis in a STING-dependent manner. Our live-cell imaging, mass spectrometry-based proteomics, CRISPR-based cellular assays, and optogenetics underscore the value of integrative approaches to uncover complex cellular responses against pathogens.
Copyright © 2016 Diner et al.
Figures

Similar articles
-
Charge-Mediated Pyrin Oligomerization Nucleates Antiviral IFI16 Sensing of Herpesvirus DNA.mBio. 2019 Jul 23;10(4):e01428-19. doi: 10.1128/mBio.01428-19. mBio. 2019. PMID: 31337724 Free PMC article.
-
The intracellular DNA sensors cGAS and IFI16 do not mediate effective antiviral immune responses to HSV-1 in human microglial cells.J Neurovirol. 2020 Aug;26(4):544-555. doi: 10.1007/s13365-020-00852-1. Epub 2020 Jun 2. J Neurovirol. 2020. PMID: 32488842 Free PMC article.
-
Interactions of the Antiviral Factor Interferon Gamma-Inducible Protein 16 (IFI16) Mediate Immune Signaling and Herpes Simplex Virus-1 Immunosuppression.Mol Cell Proteomics. 2015 Sep;14(9):2341-56. doi: 10.1074/mcp.M114.047068. Epub 2015 Feb 18. Mol Cell Proteomics. 2015. PMID: 25693804 Free PMC article.
-
Interrogating Host Antiviral Environments Driven by Nuclear DNA Sensing: A Multiomic Perspective.Biomolecules. 2020 Nov 24;10(12):1591. doi: 10.3390/biom10121591. Biomolecules. 2020. PMID: 33255247 Free PMC article. Review.
-
Nuclear sensing of viral DNA, epigenetic regulation of herpes simplex virus infection, and innate immunity.Virology. 2015 May;479-480:153-9. doi: 10.1016/j.virol.2015.02.009. Epub 2015 Mar 3. Virology. 2015. PMID: 25742715 Free PMC article. Review.
Cited by
-
Drugs Targeting Sirtuin 2 Exhibit Broad-Spectrum Anti-Infective Activity.Pharmaceuticals (Basel). 2024 Sep 29;17(10):1298. doi: 10.3390/ph17101298. Pharmaceuticals (Basel). 2024. PMID: 39458938 Free PMC article. Review.
-
The DNA Sensor cGAS is Decorated by Acetylation and Phosphorylation Modifications in the Context of Immune Signaling.Mol Cell Proteomics. 2020 Jul;19(7):1193-1208. doi: 10.1074/mcp.RA120.001981. Epub 2020 Apr 28. Mol Cell Proteomics. 2020. PMID: 32345711 Free PMC article.
-
Camouflage and interception: how pathogens evade detection by intracellular nucleic acid sensors.Immunology. 2019 Mar;156(3):217-227. doi: 10.1111/imm.13030. Epub 2018 Dec 18. Immunology. 2019. PMID: 30499584 Free PMC article. Review.
-
The HSV-1 ubiquitin ligase ICP0: Modifying the cellular proteome to promote infection.Virus Res. 2020 Aug;285:198015. doi: 10.1016/j.virusres.2020.198015. Epub 2020 May 13. Virus Res. 2020. PMID: 32416261 Free PMC article. Review.
-
Uncovering the Anticancer Potential of Murine Cytomegalovirus against Human Colon Cancer Cells.Mol Ther Oncolytics. 2020 Jan 29;16:250-261. doi: 10.1016/j.omto.2020.01.007. eCollection 2020 Mar 27. Mol Ther Oncolytics. 2020. PMID: 32140563 Free PMC article.
References
-
- Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. 2011. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe 9:363–375. doi:10.1016/j.chom.2011.04.008. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
