In vitro patterning of pluripotent stem cell-derived intestine recapitulates in vivo human development
- PMID: 27927684
- PMCID: PMC5358103
- DOI: 10.1242/dev.138453
In vitro patterning of pluripotent stem cell-derived intestine recapitulates in vivo human development
Abstract
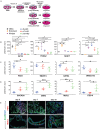
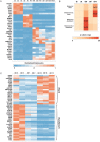
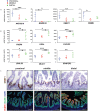
The intestine plays a central role in digestion, nutrient absorption and metabolism, with individual regions of the intestine having distinct functional roles. Many examples of region-specific gene expression in the adult intestine are known, but how intestinal regional identity is established during development is a largely unresolved issue. Here, we have identified several genes that are expressed in a region-specific manner in the developing human intestine. Using human embryonic stem cell-derived intestinal organoids, we demonstrate that the duration of exposure to active FGF and WNT signaling controls regional identity. Short-term exposure to FGF4 and CHIR99021 (a GSK3β inhibitor that stabilizes β-catenin) resulted in organoids with gene expression patterns similar to developing human duodenum, whereas longer exposure resulted in organoids similar to ileum. When region-specific organoids were transplanted into immunocompromised mice, duodenum-like organoids and ileum-like organoids retained their regional identity, demonstrating that regional identity of organoids is stable after initial patterning occurs. This work provides insights into the mechanisms that control regional specification of the developing human intestine and provides new tools for basic and translational research.
Keywords: Human; Intestine; Organoid; Patterning; Pluripotent stem cells.
© 2017. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures


Similar articles
-
Intestinal Commitment and Maturation of Human Pluripotent Stem Cells Is Independent of Exogenous FGF4 and R-spondin1.PLoS One. 2015 Jul 31;10(7):e0134551. doi: 10.1371/journal.pone.0134551. eCollection 2015. PLoS One. 2015. PMID: 26230325 Free PMC article.
-
FGF8-mediated gene regulation affects regional identity in human cerebral organoids.Elife. 2024 Nov 1;13:e98096. doi: 10.7554/eLife.98096. Elife. 2024. PMID: 39485283 Free PMC article.
-
Pluripotent stem cell differentiation reveals distinct developmental pathways regulating lung- versus thyroid-lineage specification.Development. 2017 Nov 1;144(21):3879-3893. doi: 10.1242/dev.150193. Epub 2017 Sep 25. Development. 2017. PMID: 28947536 Free PMC article.
-
A critical look: Challenges in differentiating human pluripotent stem cells into desired cell types and organoids.Wiley Interdiscip Rev Dev Biol. 2020 May;9(3):e368. doi: 10.1002/wdev.368. Epub 2019 Nov 19. Wiley Interdiscip Rev Dev Biol. 2020. PMID: 31746148 Free PMC article. Review.
-
hPSC-derived lung and intestinal organoids as models of human fetal tissue.Dev Biol. 2016 Dec 15;420(2):230-238. doi: 10.1016/j.ydbio.2016.06.006. Epub 2016 Jun 7. Dev Biol. 2016. PMID: 27287882 Free PMC article. Review.
Cited by
-
Nonadhesive Alginate Hydrogels Support Growth of Pluripotent Stem Cell-Derived Intestinal Organoids.Stem Cell Reports. 2019 Feb 12;12(2):381-394. doi: 10.1016/j.stemcr.2018.12.001. Epub 2019 Jan 3. Stem Cell Reports. 2019. PMID: 30612954 Free PMC article.
-
Generation of lung organoids from human pluripotent stem cells in vitro.Nat Protoc. 2019 Feb;14(2):518-540. doi: 10.1038/s41596-018-0104-8. Nat Protoc. 2019. PMID: 30664680 Free PMC article.
-
Morphogenesis and maturation of the embryonic and postnatal intestine.Semin Cell Dev Biol. 2017 Jun;66:81-93. doi: 10.1016/j.semcdb.2017.01.011. Epub 2017 Feb 1. Semin Cell Dev Biol. 2017. PMID: 28161556 Free PMC article. Review.
-
Elucidation of HHEX in pancreatic endoderm differentiation using a human iPSC differentiation model.Sci Rep. 2023 May 29;13(1):8659. doi: 10.1038/s41598-023-35875-1. Sci Rep. 2023. PMID: 37248264 Free PMC article.
-
Taming the Wild West of Organoids, Enteroids, and Mini-Guts.Cell Mol Gastroenterol Hepatol. 2017 Nov 28;5(2):159-160. doi: 10.1016/j.jcmgh.2017.11.003. eCollection 2018. Cell Mol Gastroenterol Hepatol. 2017. PMID: 29693042 Free PMC article. No abstract available.
References
-
- Beck C. W. and Slack J. M. (1999). A developmental pathway controlling outgrowth of the Xenopus tail bud. Development 126, 1611-1620. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 DK007762/DK/NIDDK NIH HHS/United States
- U19 AI116482/AI/NIAID NIH HHS/United States
- U24 DK085532/DK/NIDDK NIH HHS/United States
- T32 HD007505/HD/NICHD NIH HHS/United States
- U01 DK103141/DK/NIDDK NIH HHS/United States
- P30 DK034933/DK/NIDDK NIH HHS/United States
- U01 DK085532/DK/NIDDK NIH HHS/United States
- R24 HD000836/HD/NICHD NIH HHS/United States
- DP5 OD012194/OD/NIH HHS/United States
- P50 GM081879/GM/NIGMS NIH HHS/United States
- U01 DK103147/DK/NIDDK NIH HHS/United States
- T32 DK094775/DK/NIDDK NIH HHS/United States
- K01 DK091415/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

