Residue-centric modeling and design of saccharide and glycoconjugate structures
- PMID: 27900782
- PMCID: PMC5182120
- DOI: 10.1002/jcc.24679
Residue-centric modeling and design of saccharide and glycoconjugate structures
Erratum in
-
Erratum.J Comput Chem. 2017 May 15;38(13):1015. doi: 10.1002/jcc.24777. Epub 2017 Mar 16. J Comput Chem. 2017. PMID: 28340288 No abstract available.
Abstract
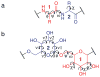
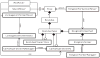
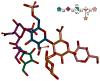
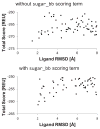
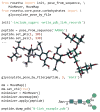
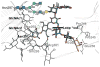
The RosettaCarbohydrate framework is a new tool for modeling a wide variety of saccharide and glycoconjugate structures. This report describes the development of the framework and highlights its applications. The framework integrates with established protocols within the Rosetta modeling and design suite, and it handles the vast complexity and variety of carbohydrate molecules, including branching and sugar modifications. To address challenges of sampling and scoring, RosettaCarbohydrate can sample glycosidic bonds, side-chain conformations, and ring forms, and it utilizes a glycan-specific term within its scoring function. Rosetta can work with standard PDB, GLYCAM, and GlycoWorkbench (.gws) file formats. Saccharide residue-specific chemical information is stored internally, permitting glycoengineering and design. Carbohydrate-specific applications described herein include virtual glycosylation, loop-modeling of carbohydrates, and docking of glyco-ligands to antibodies. Benchmarking data are presented and compared to other studies, demonstrating Rosetta's ability to predict glyco-ligand binding. The framework expands the tools available to glycoscientists and engineers. © 2016 Wiley Periodicals, Inc.
Keywords: Rosetta; carbohydrates; computation; docking; glycoconjugates; glycoproteins; modeling; oligosaccharides; structure.
© 2016 Wiley Periodicals, Inc.
Figures

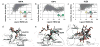
Similar articles
-
CHARMM-GUI Glycan Modeler for modeling and simulation of carbohydrates and glycoconjugates.Glycobiology. 2019 Apr 1;29(4):320-331. doi: 10.1093/glycob/cwz003. Glycobiology. 2019. PMID: 30689864 Free PMC article.
-
A perspective on the primary and three-dimensional structures of carbohydrates.Carbohydr Res. 2013 Aug 30;378:123-32. doi: 10.1016/j.carres.2013.02.005. Epub 2013 Feb 24. Carbohydr Res. 2013. PMID: 23522728 Review.
-
Glycosylator: a Python framework for the rapid modeling of glycans.BMC Bioinformatics. 2019 Oct 22;20(1):513. doi: 10.1186/s12859-019-3097-6. BMC Bioinformatics. 2019. PMID: 31640540 Free PMC article.
-
Strategies for glycoconjugate analysis.Acta Anat (Basel). 1998;161(1-4):18-35. doi: 10.1159/000046449. Acta Anat (Basel). 1998. PMID: 9780350
-
Interfering with the sugar code: design and synthesis of oligosaccharide mimics.Chemistry. 2008;14(25):7434-41. doi: 10.1002/chem.200800597. Chemistry. 2008. PMID: 18613175 Review.
Cited by
-
In Vivo Assembly of Nanoparticles Achieved through Synergy of Structure-Based Protein Engineering and Synthetic DNA Generates Enhanced Adaptive Immunity.Adv Sci (Weinh). 2020 Feb 27;7(8):1902802. doi: 10.1002/advs.201902802. eCollection 2020 Apr. Adv Sci (Weinh). 2020. PMID: 32328416 Free PMC article.
-
The Rosetta All-Atom Energy Function for Macromolecular Modeling and Design.J Chem Theory Comput. 2017 Jun 13;13(6):3031-3048. doi: 10.1021/acs.jctc.7b00125. Epub 2017 May 12. J Chem Theory Comput. 2017. PMID: 28430426 Free PMC article.
-
Computational tools for drawing, building and displaying carbohydrates: a visual guide.Beilstein J Org Chem. 2020 Oct 2;16:2448-2468. doi: 10.3762/bjoc.16.199. eCollection 2020. Beilstein J Org Chem. 2020. PMID: 33082879 Free PMC article. Review.
-
Development and Evaluation of GlycanDock: A Protein-Glycoligand Docking Refinement Algorithm in Rosetta.J Phys Chem B. 2021 Jun 16:10.1021/acs.jpcb.1c00910. doi: 10.1021/acs.jpcb.1c00910. Online ahead of print. J Phys Chem B. 2021. PMID: 34133179 Free PMC article.
-
CHARMM-GUI Glycan Modeler for modeling and simulation of carbohydrates and glycoconjugates.Glycobiology. 2019 Apr 1;29(4):320-331. doi: 10.1093/glycob/cwz003. Glycobiology. 2019. PMID: 30689864 Free PMC article.
References
-
- Committee on Assessing the Importance and Impact of Glycomics and Glycosciences, Board on Chemical Sciences and Technology, Board on Life Sciences, Division on Earth and Life Studies, National Research Council. Transforming Glycoscience: A Roadmap for the Future. National Academies Press; Washington, DC: 2012. - PubMed
-
- Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. Essentials of Glycobiology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2008. - PubMed
-
- Werz DB, Ranzinger R, Herget S, Adibekian A, von der Lieth CW, Seeberger PH. ACS Chem Biol. 2007;2:685–691. - PubMed
-
- Krambeck FJ, Betenbaugh MJ. Biotechnol Bioeng. 2005;92:711–728. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

