Different enhancer classes in Drosophila bind distinct architectural proteins and mediate unique chromatin interactions and 3D architecture
- PMID: 27899590
- PMCID: PMC5389536
- DOI: 10.1093/nar/gkw1114
Different enhancer classes in Drosophila bind distinct architectural proteins and mediate unique chromatin interactions and 3D architecture
Abstract
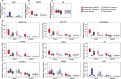
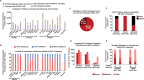
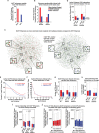
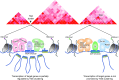
Eukaryotic gene expression is regulated by enhancer-promoter interactions but the molecular mechanisms that govern specificity have remained elusive. Genome-wide studies utilizing STARR-seq identified two enhancer classes in Drosophila that interact with different core promoters: housekeeping enhancers (hkCP) and developmental enhancers (dCP). We hypothesized that the two enhancer classes are occupied by distinct architectural proteins, affecting their enhancer-promoter contacts. By evaluating ChIP-seq occupancy of architectural proteins, typical enhancer-associated proteins, and histone modifications, we determine that both enhancer classes are enriched for RNA Polymerase II, CBP, and architectural proteins but there are also distinctions. hkCP enhancers contain H3K4me3 and exclusively bind Cap-H2, Chromator, DREF and Z4, whereas dCP enhancers contain H3K4me1 and are more enriched for Rad21 and Fs(1)h-L. Additionally, we map the interactions of each enhancer class utilizing a Hi-C dataset with <1 kb resolution. Results suggest that hkCP enhancers are more likely to form multi-TSS interaction networks and be associated with topologically associating domain (TAD) borders, while dCP enhancers are more often bound to one or two TSSs and are enriched at chromatin loop anchors. The data support a model suggesting that the unique architectural protein occupancy within enhancers is one contributor to enhancer-promoter interaction specificity.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
5C analysis of the Epidermal Differentiation Complex locus reveals distinct chromatin interaction networks between gene-rich and gene-poor TADs in skin epithelial cells.PLoS Genet. 2017 Sep 1;13(9):e1006966. doi: 10.1371/journal.pgen.1006966. eCollection 2017 Sep. PLoS Genet. 2017. PMID: 28863138 Free PMC article.
-
Genome-wide phosphoacetylation of histone H3 at Drosophila enhancers and promoters.Genome Res. 2012 Jun;22(6):1081-8. doi: 10.1101/gr.136929.111. Epub 2012 Apr 16. Genome Res. 2012. PMID: 22508764 Free PMC article.
-
DNA methylation regulates discrimination of enhancers from promoters through a H3K4me1-H3K4me3 seesaw mechanism.BMC Genomics. 2017 Dec 12;18(1):964. doi: 10.1186/s12864-017-4353-7. BMC Genomics. 2017. PMID: 29233090 Free PMC article.
-
Chromatin insulators and long-distance interactions in Drosophila.FEBS Lett. 2014 Jan 3;588(1):8-14. doi: 10.1016/j.febslet.2013.10.039. Epub 2013 Nov 5. FEBS Lett. 2014. PMID: 24211836 Review.
-
Evaluating Enhancer Function and Transcription.Annu Rev Biochem. 2020 Jun 20;89:213-234. doi: 10.1146/annurev-biochem-011420-095916. Epub 2020 Mar 20. Annu Rev Biochem. 2020. PMID: 32197056 Review.
Cited by
-
Forecasting histone methylation by Polycomb complexes with minute-scale precision.Sci Adv. 2023 Dec 22;9(51):eadj8198. doi: 10.1126/sciadv.adj8198. Epub 2023 Dec 22. Sci Adv. 2023. PMID: 38134278 Free PMC article.
-
The Insulator Protein CTCF Is Required for Correct Hox Gene Expression, but Not for Embryonic Development in Drosophila.Genetics. 2018 Sep;210(1):129-136. doi: 10.1534/genetics.118.301350. Epub 2018 Jul 18. Genetics. 2018. PMID: 30021792 Free PMC article.
-
Damage-responsive elements in Drosophila regeneration.Genome Res. 2018 Dec;28(12):1852-1866. doi: 10.1101/gr.233098.117. Epub 2018 Nov 20. Genome Res. 2018. PMID: 30459214 Free PMC article.
-
Chromosome organization in bacteria: mechanistic insights into genome structure and function.Nat Rev Genet. 2020 Apr;21(4):227-242. doi: 10.1038/s41576-019-0185-4. Epub 2019 Nov 25. Nat Rev Genet. 2020. PMID: 31767998 Review.
-
Single-cell absolute contact probability detection reveals chromosomes are organized by multiple low-frequency yet specific interactions.Nat Commun. 2017 Nov 24;8(1):1753. doi: 10.1038/s41467-017-01962-x. Nat Commun. 2017. PMID: 29170434 Free PMC article.
References
-
- Vernimmen D., Bickmore W.A.. The hierarchy of transcriptional activation: from enhancer to promoter. Trends Genet. 2015; 31:696–708. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

