Glutaminolysis and Fumarate Accumulation Integrate Immunometabolic and Epigenetic Programs in Trained Immunity
- PMID: 27866838
- PMCID: PMC5742541
- DOI: 10.1016/j.cmet.2016.10.008
Glutaminolysis and Fumarate Accumulation Integrate Immunometabolic and Epigenetic Programs in Trained Immunity
Abstract
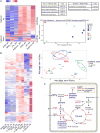
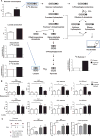
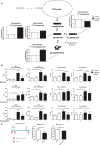
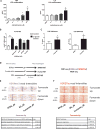
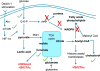
Induction of trained immunity (innate immune memory) is mediated by activation of immune and metabolic pathways that result in epigenetic rewiring of cellular functional programs. Through network-level integration of transcriptomics and metabolomics data, we identify glycolysis, glutaminolysis, and the cholesterol synthesis pathway as indispensable for the induction of trained immunity by β-glucan in monocytes. Accumulation of fumarate, due to glutamine replenishment of the TCA cycle, integrates immune and metabolic circuits to induce monocyte epigenetic reprogramming by inhibiting KDM5 histone demethylases. Furthermore, fumarate itself induced an epigenetic program similar to β-glucan-induced trained immunity. In line with this, inhibition of glutaminolysis and cholesterol synthesis in mice reduced the induction of trained immunity by β-glucan. Identification of the metabolic pathways leading to induction of trained immunity contributes to our understanding of innate immune memory and opens new therapeutic avenues.
Keywords: cholesterol metabolism; epigenetics; glutamine metabolism; glycolysis; trained immunity.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
Similar articles
-
miR-9-5p regulates immunometabolic and epigenetic pathways in β-glucan-trained immunity via IDH3α.JCI Insight. 2021 May 10;6(9):e144260. doi: 10.1172/jci.insight.144260. JCI Insight. 2021. PMID: 33986196 Free PMC article.
-
mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity.Science. 2014 Sep 26;345(6204):1250684. doi: 10.1126/science.1250684. Science. 2014. PMID: 25258083 Free PMC article.
-
Immunometabolic circuits in trained immunity.Semin Immunol. 2016 Oct;28(5):425-430. doi: 10.1016/j.smim.2016.09.002. Epub 2016 Sep 27. Semin Immunol. 2016. PMID: 27686054 Review.
-
Immunometabolic control of trained immunity.Mol Aspects Med. 2021 Feb;77:100897. doi: 10.1016/j.mam.2020.100897. Epub 2020 Sep 2. Mol Aspects Med. 2021. PMID: 32891423 Free PMC article. Review.
-
Immunometabolic changes of β-glucan-trained immunity induction and inhibition on neonatal calf immune innate cells.Mol Immunol. 2023 Jul;159:58-68. doi: 10.1016/j.molimm.2023.05.008. Epub 2023 Jun 2. Mol Immunol. 2023. PMID: 37271010
Cited by
-
Glutamine metabolism in adipocytes: a bona fide epigenetic modulator of inflammation.Adipocyte. 2020 Dec;9(1):620-625. doi: 10.1080/21623945.2020.1831825. Adipocyte. 2020. PMID: 33043853 Free PMC article. Review.
-
Overcoming immune dysfunction in the elderly: trained immunity as a novel approach.Int Immunol. 2020 Nov 23;32(12):741-753. doi: 10.1093/intimm/dxaa052. Int Immunol. 2020. PMID: 32766848 Free PMC article. Review.
-
miR-9-5p regulates immunometabolic and epigenetic pathways in β-glucan-trained immunity via IDH3α.JCI Insight. 2021 May 10;6(9):e144260. doi: 10.1172/jci.insight.144260. JCI Insight. 2021. PMID: 33986196 Free PMC article.
-
The Role of the Pathogen Dose and PI3Kγ in Immunometabolic Reprogramming of Microglia for Innate Immune Memory.Int J Mol Sci. 2021 Mar 4;22(5):2578. doi: 10.3390/ijms22052578. Int J Mol Sci. 2021. PMID: 33806610 Free PMC article.
-
Biofilm and Small Colony Variants-An Update on Staphylococcus aureus Strategies toward Drug Resistance.Int J Mol Sci. 2022 Jan 22;23(3):1241. doi: 10.3390/ijms23031241. Int J Mol Sci. 2022. PMID: 35163165 Free PMC article. Review.
References
-
- Chen Q, Giedt M, Tang L, Harrison DA. Tools and methods for studying the Drosophila JAK/STAT pathway. Methods. 2014;68:160–172. - PubMed
-
- Cheng SC, Scicluna BP, Arts RJ, Gresnigt MS, Lachmandas E, Giamarellos-Bourboulis EJ, Kox M, Manjeri GR, Wagenaars JA, Cremer OL, et al. Broad defects in the energy metabolism of leukocytes underlie immunoparalysis in sepsis. Nat Immunol. 2016;17:406–413. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

