Structure of the NS2B-NS3 protease from Zika virus after self-cleavage
- PMID: 27845325
- PMCID: PMC5116066
- DOI: 10.1038/ncomms13410
Structure of the NS2B-NS3 protease from Zika virus after self-cleavage
Abstract
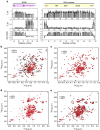
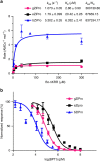
The recent outbreak of Zika virus (ZIKV) infections in the Americas represents a serious threat to the global public health. The viral protease that processes viral polyproteins during infection appears as an attractive drug target. Here we report a crystal structure at 1.84 Å resolution of ZIKV non-structural protein NS2B-NS3 protease with the last four amino acids of the NS2B cofactor bound at the NS3 active site. This structure represents a post-proteolysis state of the enzyme during viral polyprotein processing and provides insights into peptide substrate recognition by the protease. Nuclear magnetic resonance (NMR) studies and protease activity assays unravel the protein dynamics upon binding the protease inhibitor BPTI in solution and confirm this finding. The structural and functional insights of the ZIKV protease presented here should advance our current understanding of flavivirus replication and accelerate structure-based antiviral drug discovery against ZIKV.
Figures

Similar articles
-
Crystal structure of unlinked NS2B-NS3 protease from Zika virus.Science. 2016 Dec 23;354(6319):1597-1600. doi: 10.1126/science.aai9309. Epub 2016 Dec 8. Science. 2016. PMID: 27940580
-
Characterization of molecular interactions between Zika virus protease and peptides derived from the C-terminus of NS2B.Biochem Biophys Res Commun. 2018 Sep 5;503(2):691-696. doi: 10.1016/j.bbrc.2018.06.062. Epub 2018 Jun 21. Biochem Biophys Res Commun. 2018. PMID: 29908184
-
Dynamic Interactions of Post Cleaved NS2B Cofactor and NS3 Protease Identified by Integrative Structural Approaches.Viruses. 2022 Jun 30;14(7):1440. doi: 10.3390/v14071440. Viruses. 2022. PMID: 35891424 Free PMC article.
-
The Structure of the Zika Virus Protease, NS2B/NS3pro.Adv Exp Med Biol. 2018;1062:131-145. doi: 10.1007/978-981-10-8727-1_10. Adv Exp Med Biol. 2018. PMID: 29845530 Review.
-
Zika Virus Protease: An Antiviral Drug Target.Trends Microbiol. 2017 Oct;25(10):797-808. doi: 10.1016/j.tim.2017.07.001. Epub 2017 Aug 5. Trends Microbiol. 2017. PMID: 28789826 Review.
Cited by
-
Quantum Biochemistry and MM-PBSA Description of the ZIKV NS2B-NS3 Protease: Insights into the Binding Interactions beyond the Catalytic Triad Pocket.Int J Mol Sci. 2022 Sep 3;23(17):10088. doi: 10.3390/ijms231710088. Int J Mol Sci. 2022. PMID: 36077486 Free PMC article.
-
Development of Small-Molecule Inhibitors Against Zika Virus Infection.Front Microbiol. 2019 Dec 6;10:2725. doi: 10.3389/fmicb.2019.02725. eCollection 2019. Front Microbiol. 2019. PMID: 31866959 Free PMC article. Review.
-
Chalcones from Angelica keiskei (ashitaba) inhibit key Zika virus replication proteins.Bioorg Chem. 2022 Mar;120:105649. doi: 10.1016/j.bioorg.2022.105649. Epub 2022 Jan 31. Bioorg Chem. 2022. PMID: 35124513 Free PMC article.
-
De Novo Discovery of Nonstandard Macrocyclic Peptides as Noncompetitive Inhibitors of the Zika Virus NS2B-NS3 Protease.ACS Med Chem Lett. 2019 Jan 4;10(2):168-174. doi: 10.1021/acsmedchemlett.8b00535. eCollection 2019 Feb 14. ACS Med Chem Lett. 2019. PMID: 30783498 Free PMC article.
-
Zika virus NS3 protease induces bone morphogenetic protein-dependent brain calcification in human fetuses.Nat Microbiol. 2021 Apr;6(4):455-466. doi: 10.1038/s41564-020-00850-3. Epub 2021 Jan 28. Nat Microbiol. 2021. PMID: 33510473 Free PMC article.
References
-
- Petersen L. R., Jamieson D. J., Powers A. M. & Honein M. A. Zika virus. N. Engl. J. Med. 374, 1552–1563 (2016). - PubMed
-
- Broutet N. et al.. Zika virus as a cause of neurologic disorders. N. Engl. J. Med. 374, 1506–1509 (2016). - PubMed
-
- Carteaux G. et al.. Zika virus associated with meningoencephalitis. N. Engl. J. Med. 374, 1595–1596 (2016). - PubMed
-
- Mecharles S. et al.. Acute myelitis due to Zika virus infection. Lancet 387, 1481 (2016). - PubMed
-
- Pierson T. C. & Diamond M. S. in Fields Virology 6th edn. Ch. 26, 747–794 (Lippincott-Raven Publishers, 2013).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

