Macrophage Ontogeny Underlies Differences in Tumor-Specific Education in Brain Malignancies
- PMID: 27840052
- PMCID: PMC5450644
- DOI: 10.1016/j.celrep.2016.10.052
Macrophage Ontogeny Underlies Differences in Tumor-Specific Education in Brain Malignancies
Abstract
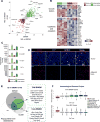
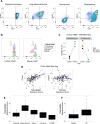
Extensive transcriptional and ontogenetic diversity exists among normal tissue-resident macrophages, with unique transcriptional profiles endowing the cells with tissue-specific functions. However, it is unknown whether the origins of different macrophage populations affect their roles in malignancy. Given potential artifacts associated with irradiation-based lineage tracing, it remains unclear if bone-marrow-derived macrophages (BMDMs) are present in tumors of the brain, a tissue with no homeostatic involvement of BMDMs. Here, we employed multiple models of murine brain malignancy and genetic lineage tracing to demonstrate that BMDMs are abundant in primary and metastatic brain tumors. Our data indicate that distinct transcriptional networks in brain-resident microglia and recruited BMDMs are associated with tumor-mediated education yet are also influenced by chromatin landscapes established before tumor initiation. Furthermore, we demonstrate that microglia specifically repress Itga4 (CD49D), enabling its utility as a discriminatory marker between microglia and BMDMs in primary and metastatic disease in mouse and human.
Keywords: CD49D; Macrophage; brain metastasis; glioma; microglia; tumor-associated macrophages.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Distinct roles of CSF family cytokines in macrophage infiltration and activation in glioma progression and injury response.J Pathol. 2013 Jul;230(3):310-21. doi: 10.1002/path.4192. J Pathol. 2013. PMID: 23520016
-
Resident microglia, and not peripheral macrophages, are the main source of brain tumor mononuclear cells.Int J Cancer. 2015 Jul 15;137(2):278-88. doi: 10.1002/ijc.29379. Epub 2014 Dec 24. Int J Cancer. 2015. PMID: 25477239
-
Deletion of Neuropilin 1 from Microglia or Bone Marrow-Derived Macrophages Slows Glioma Progression.Cancer Res. 2018 Feb 1;78(3):685-694. doi: 10.1158/0008-5472.CAN-17-1435. Epub 2017 Nov 2. Cancer Res. 2018. PMID: 29097606 Free PMC article.
-
From bone marrow to microglia: barriers and avenues.Trends Immunol. 2008 May;29(5):227-34. doi: 10.1016/j.it.2008.01.010. Epub 2008 Apr 7. Trends Immunol. 2008. PMID: 18396103 Review.
-
Bone marrow-derived macrophages and the CNS: An update on the use of experimental chimeric mouse models and bone marrow transplantation in neurological disorders.Biochim Biophys Acta. 2016 Mar;1862(3):310-22. doi: 10.1016/j.bbadis.2015.09.017. Epub 2015 Oct 8. Biochim Biophys Acta. 2016. PMID: 26432480 Review.
Cited by
-
Analyzing One Cell at a TIME: Analysis of Myeloid Cell Contributions in the Tumor Immune Microenvironment.Front Immunol. 2020 Sep 2;11:1842. doi: 10.3389/fimmu.2020.01842. eCollection 2020. Front Immunol. 2020. PMID: 32983100 Free PMC article. Review.
-
Macrophages in melanoma: A double‑edged sword and targeted therapy strategies (Review).Exp Ther Med. 2022 Aug 26;24(4):640. doi: 10.3892/etm.2022.11577. eCollection 2022 Oct. Exp Ther Med. 2022. PMID: 36160877 Free PMC article. Review.
-
Pancancer landscape analysis of the thymosin family identified TMSB10 as a potential prognostic biomarker and immunotherapy target in glioma.Cancer Cell Int. 2022 Sep 26;22(1):294. doi: 10.1186/s12935-022-02698-5. Cancer Cell Int. 2022. PMID: 36163046 Free PMC article.
-
PVT1 promotes proliferation and macrophage immunosuppressive polarization through STAT1 and CX3CL1 regulation in glioblastoma multiforme.CNS Neurosci Ther. 2024 Jan;30(1):e14566. doi: 10.1111/cns.14566. CNS Neurosci Ther. 2024. PMID: 38287522 Free PMC article.
-
Self-Renewal of Macrophages: Tumor-Released Factors and Signaling Pathways.Biomedicines. 2022 Oct 26;10(11):2709. doi: 10.3390/biomedicines10112709. Biomedicines. 2022. PMID: 36359228 Free PMC article. Review.
References
-
- Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FM. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat Neurosci. 2007;10:1538–1543. - PubMed
-
- Biffi A, De Palma M, Quattrini A, Del Carro U, Amadio S, Visigalli I, Sessa M, Fasano S, Brambilla R, Marchesini S, et al. Correction of metachromatic leukodystrophy in the mouse model by transplantation of genetically modified hematopoietic stem cells. J Clin Invest. 2004;113:1118–1129. - PMC - PubMed
-
- Bowman RL, Joyce JA. Therapeutic targeting of tumor-associated macrophages and microglia in glioblastoma. Immunotherapy. 2014;6:663–666. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

