The RNA methyltransferase Dnmt2 methylates DNA in the structural context of a tRNA
- PMID: 27819523
- PMCID: PMC5699543
- DOI: 10.1080/15476286.2016.1236170
The RNA methyltransferase Dnmt2 methylates DNA in the structural context of a tRNA
Abstract
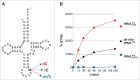
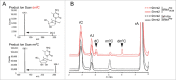
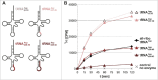
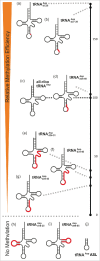
The amino acid sequence of Dnmt2 is very similar to the catalytic domains of bacterial and eukaryotic DNA-(cytosine 5)-methyltransferases, but it efficiently catalyzes tRNA methylation, while its DNA methyltransferase activity is the subject of controversial reports with rates varying between zero and very weak. By using composite nucleic acid molecules as substrates, we surprisingly found that DNA fragments, when presented as covalent DNA-RNA hybrids in the structural context of a tRNA, can be more efficiently methylated than the corresponding natural tRNA substrate. Furthermore, by stepwise development of tRNAAsp, we showed that this natural Dnmt2 substrate could be engineered to employ RNAs that act like guide RNAs in vitro. The 5'-half of tRNAAsp was able to efficiently guide methylation toward a single stranded tRNA fragment as would result from tRNA cleavage by tRNA specific nucleases. In a more artificial setting, a composite system of guide RNAs could ultimately be engineered to enable the enzyme to perform cytidine methylation on single stranded DNA in vitro.
Keywords: 5-methylcytosine; DNA; Dnmt2; RNA methylation; RNA modification; enzyme kinetics; modification pathway crosstalk; tRNA.
Figures


Similar articles
-
New substrates and determinants for tRNA recognition of RNA methyltransferase DNMT2/TRDMT1.RNA Biol. 2021 Dec;18(12):2531-2545. doi: 10.1080/15476286.2021.1930756. Epub 2021 Jun 10. RNA Biol. 2021. PMID: 34110975 Free PMC article.
-
RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage.Genes Dev. 2010 Aug 1;24(15):1590-5. doi: 10.1101/gad.586710. Genes Dev. 2010. PMID: 20679393 Free PMC article.
-
Human DNMT2 methylates tRNA(Asp) molecules using a DNA methyltransferase-like catalytic mechanism.RNA. 2008 Aug;14(8):1663-70. doi: 10.1261/rna.970408. Epub 2008 Jun 20. RNA. 2008. PMID: 18567810 Free PMC article.
-
Cross-Talk between Dnmt2-Dependent tRNA Methylation and Queuosine Modification.Biomolecules. 2017 Feb 10;7(1):14. doi: 10.3390/biom7010014. Biomolecules. 2017. PMID: 28208632 Free PMC article. Review.
-
Mechanism and biological role of Dnmt2 in Nucleic Acid Methylation.RNA Biol. 2017 Sep 2;14(9):1108-1123. doi: 10.1080/15476286.2016.1191737. Epub 2016 May 27. RNA Biol. 2017. PMID: 27232191 Free PMC article. Review.
Cited by
-
Sleep Deprivation and the Epigenome.Front Neural Circuits. 2018 Feb 27;12:14. doi: 10.3389/fncir.2018.00014. eCollection 2018. Front Neural Circuits. 2018. PMID: 29535611 Free PMC article. Review.
-
Neuronal Nsun2 deficiency produces tRNA epitranscriptomic alterations and proteomic shifts impacting synaptic signaling and behavior.Nat Commun. 2021 Aug 13;12(1):4913. doi: 10.1038/s41467-021-24969-x. Nat Commun. 2021. PMID: 34389722 Free PMC article.
-
The Dictyostelium discoideum genome lacks significant DNA methylation and uncovers palindromic sequences as a source of false positives in bisulfite sequencing.NAR Genom Bioinform. 2023 Apr 18;5(2):lqad035. doi: 10.1093/nargab/lqad035. eCollection 2023 Jun. NAR Genom Bioinform. 2023. PMID: 37081864 Free PMC article.
-
Epigenetics in blood-brain barrier disruption.Fluids Barriers CNS. 2021 Apr 6;18(1):17. doi: 10.1186/s12987-021-00250-7. Fluids Barriers CNS. 2021. PMID: 33823899 Free PMC article. Review.
-
Head and Neck Squamous Cell Carcinoma: Epigenetic Landscape.Diagnostics (Basel). 2020 Dec 27;11(1):34. doi: 10.3390/diagnostics11010034. Diagnostics (Basel). 2020. PMID: 33375464 Free PMC article. Review.
References
-
- Rubio MA, Pastar I, Gaston KW, Ragone FL, Janzen CJ, Cross GA, Papavasiliou FN, Alfonzo JD. An adenosine-to-inosine tRNA-editing enzyme that can perform C-to-U deamination of DNA. Proc Natl Acad Sci U S A 2007; 104:7821–6; PMID:17483465; http://dx.doi.org/10.1073/pnas.0702394104 - DOI - PMC - PubMed
-
- Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell 2002; 10:1247–53; PMID:12453430; http://dx.doi.org/10.1016/S1097-2765(02)00742-6 - DOI - PubMed
-
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 2006; 311:395–8; PMID:16424344; http://dx.doi.org/10.1126/science.1120976 - DOI - PubMed
-
- Jeltsch A, Ehrenhofer-Murray A, Jurkowski T, Lyko F, Reuter G, Ankri S, Nellen W, Schaefer M, Helm M. Mechanism and biological role of Dnmt2 in nucleic acid methylation. RNA Biol 2016:0; PMID:27232191; http://dx.doi.org/10.1080/15476286.2016.1191737 - DOI - PMC - PubMed
-
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 2010; 24:1590–5; PMID:20679393; http://dx.doi.org/10.1101/gad.586710 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
