The human papillomavirus E7 oncoprotein as a regulator of transcription
- PMID: 27818212
- PMCID: PMC5325776
- DOI: 10.1016/j.virusres.2016.10.017
The human papillomavirus E7 oncoprotein as a regulator of transcription
Abstract
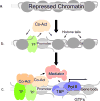
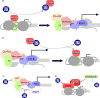
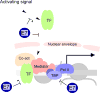
High-risk human papillomaviruses (HPVs) encode oncoproteins which manipulate gene expression patterns in the host keratinocytes to facilitate viral replication, regulate viral transcription, and promote immune evasion and persistence. In some cases, oncoprotein-induced changes in host cell behavior can cause progression to cancer, but a complete picture of the functions of the viral oncoproteins in the productive HPV life cycle remains elusive. E7 is the HPV-encoded factor most responsible for maintaining cell cycle competence in differentiating keratinocytes. Through interactions with dozens of host factors, E7 has an enormous impact on host gene expression patterns. In this review, we will examine the role of E7 specifically as a regulator of transcription. We will discuss mechanisms of regulation of cell cycle-related genes by E7 as well as genes involved in immune regulation, growth factor signaling, DNA damage responses, microRNAs, and others pathways. We will also discuss some unanswered questions about how transcriptional regulation by E7 impacts the biology of HPV in both benign and malignant conditions.
Keywords: Cell cycle control; E7; Histone deacetylase; Innate immunity; Transcription factor; pRb family.
Copyright © 2016 Elsevier B.V. All rights reserved.
Figures


Similar articles
-
Activation of telomerase by HPVs.Virus Res. 2017 Mar 2;231:50-55. doi: 10.1016/j.virusres.2016.11.003. Epub 2016 Nov 15. Virus Res. 2017. PMID: 27863966 Review.
-
Evasion of host immune defenses by human papillomavirus.Virus Res. 2017 Mar 2;231:21-33. doi: 10.1016/j.virusres.2016.11.023. Epub 2016 Nov 24. Virus Res. 2017. PMID: 27890631 Free PMC article. Review.
-
Human Papillomavirus E7 Oncoprotein Subverts Host Innate Immunity via SUV39H1-Mediated Epigenetic Silencing of Immune Sensor Genes.J Virol. 2020 Jan 31;94(4):e01812-19. doi: 10.1128/JVI.01812-19. Print 2020 Jan 31. J Virol. 2020. PMID: 31776268 Free PMC article.
-
Involvement of Brd4 in different steps of the papillomavirus life cycle.Virus Res. 2017 Mar 2;231:76-82. doi: 10.1016/j.virusres.2016.12.006. Epub 2016 Dec 10. Virus Res. 2017. PMID: 27965149 Free PMC article. Review.
-
Human papillomavirus E7 oncoprotein increases production of the anti-inflammatory interleukin-18 binding protein in keratinocytes.J Virol. 2014 Apr;88(8):4173-9. doi: 10.1128/JVI.02546-13. Epub 2014 Jan 29. J Virol. 2014. PMID: 24478434 Free PMC article.
Cited by
-
The role of HR-HPV integration in the progression of premalignant lesions into different cancer types.Heliyon. 2024 Jul 22;10(15):e34999. doi: 10.1016/j.heliyon.2024.e34999. eCollection 2024 Aug 15. Heliyon. 2024. PMID: 39170128 Free PMC article. Review.
-
Characterization of the High-Affinity Fuzzy Complex between the Disordered Domain of the E7 Oncoprotein from High-Risk HPV and the TAZ2 Domain of CBP.Biochemistry. 2021 Dec 28;60(51):3887-3898. doi: 10.1021/acs.biochem.1c00669. Epub 2021 Dec 14. Biochemistry. 2021. PMID: 34905914 Free PMC article.
-
Management of HPV-Related Squamous Cell Carcinoma of the Head and Neck: Pitfalls and Caveat.Cancers (Basel). 2020 Apr 15;12(4):975. doi: 10.3390/cancers12040975. Cancers (Basel). 2020. PMID: 32326465 Free PMC article. Review.
-
Targeted regulation of miR-154-5p/Cullin2 pathway by hsa_circ_TRIM22 in promoting human papillomavirus 16 positive cervical cancer progression.J Cancer. 2024 Feb 24;15(8):2137-2146. doi: 10.7150/jca.92631. eCollection 2024. J Cancer. 2024. PMID: 38495497 Free PMC article.
-
It Takes Two to Tango: A Review of Oncogenic Virus and Host Microbiome Associated Inflammation in Head and Neck Cancer.Cancers (Basel). 2022 Jun 25;14(13):3120. doi: 10.3390/cancers14133120. Cancers (Basel). 2022. PMID: 35804891 Free PMC article. Review.
References
-
- Alcocer-Gonzalez JM, Berumen J, et al. In vivo expression of immunosuppressive cytokines in human papillomavirus-transformed cervical cancer cells. Viral Immunol. 2006;19(3):481–491. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

