Kaumoebavirus, a New Virus That Clusters with Faustoviruses and Asfarviridae
- PMID: 27801826
- PMCID: PMC5127008
- DOI: 10.3390/v8110278
Kaumoebavirus, a New Virus That Clusters with Faustoviruses and Asfarviridae
Abstract
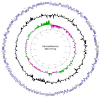
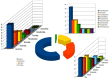
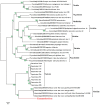
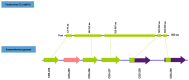
In this study, we report the isolation of a new giant virus found in sewage water from the southern area of Jeddah (Saudi Arabia), with morphological and genomic resemblance to Faustoviruses. This new giant virus, named Kaumoebavirus, was obtained from co-culture with Vermamoeba vermiformis, an amoeboid protozoa considered to be of special interest to human health and the environment. This new virus has ~250 nm icosahedral capsids and a 350,731 bp DNA genome length. The genome of Kaumoebavirus has a coding density of 86%, corresponding to 465 genes. Most of these genes (59%) are closely related to genes from members of the proposed order Megavirales, and the best matches to its proteins with other members of the Megavirales are Faustoviruses (43%) and Asfarviruses (23%). Unsurprisingly, phylogenetic reconstruction places Kaumoebavirus as a distant relative of Faustoviruses and Asfarviruses.
Keywords: Asfarviruses; Faustoviruses; Kaumoebavirus; Vermamoeba vermiformis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
The Kaumoebavirus LCC10 Genome Reveals a Unique Gene Strand Bias among "Extended Asfarviridae".Viruses. 2021 Jan 20;13(2):148. doi: 10.3390/v13020148. Viruses. 2021. PMID: 33498382 Free PMC article.
-
Pacmanvirus, a New Giant Icosahedral Virus at the Crossroads between Asfarviridae and Faustoviruses.J Virol. 2017 Jun 26;91(14):e00212-17. doi: 10.1128/JVI.00212-17. Print 2017 Jul 15. J Virol. 2017. PMID: 28446673 Free PMC article.
-
Faustovirus, an asfarvirus-related new lineage of giant viruses infecting amoebae.J Virol. 2015 Jul;89(13):6585-94. doi: 10.1128/JVI.00115-15. J Virol. 2015. PMID: 25878099 Free PMC article.
-
Asfarviruses and Closely Related Giant Viruses.Viruses. 2023 Apr 20;15(4):1015. doi: 10.3390/v15041015. Viruses. 2023. PMID: 37112995 Free PMC article. Review.
-
Giant Viruses of Amoebae: A Journey Through Innovative Research and Paradigm Changes.Annu Rev Virol. 2017 Sep 29;4(1):61-85. doi: 10.1146/annurev-virology-101416-041816. Epub 2017 Jul 31. Annu Rev Virol. 2017. PMID: 28759330 Review.
Cited by
-
Diversification of giant and large eukaryotic dsDNA viruses predated the origin of modern eukaryotes.Proc Natl Acad Sci U S A. 2019 Sep 24;116(39):19585-19592. doi: 10.1073/pnas.1912006116. Epub 2019 Sep 10. Proc Natl Acad Sci U S A. 2019. PMID: 31506349 Free PMC article.
-
Vermamoeba vermiformis: a Free-Living Amoeba of Interest.Microb Ecol. 2018 Nov;76(4):991-1001. doi: 10.1007/s00248-018-1199-8. Epub 2018 May 8. Microb Ecol. 2018. PMID: 29737382 Review.
-
Tupanvirus-infected amoebas are induced to aggregate with uninfected cells promoting viral dissemination.Sci Rep. 2019 Jan 17;9(1):183. doi: 10.1038/s41598-018-36552-4. Sci Rep. 2019. PMID: 30655573 Free PMC article.
-
The consequences of viral infection on protists.Commun Biol. 2024 Mar 11;7(1):306. doi: 10.1038/s42003-024-06001-2. Commun Biol. 2024. PMID: 38462656 Free PMC article. Review.
-
Discovery and Further Studies on Giant Viruses at the IHU Mediterranee Infection That Modified the Perception of the Virosphere.Viruses. 2019 Mar 30;11(4):312. doi: 10.3390/v11040312. Viruses. 2019. PMID: 30935049 Free PMC article. Review.
References
-
- Boyer M., Yutin N., Pagnier I., Barrassi L., Fournous G., Espinosa L., Robert C., Azza S., Sun S., Rossmann M.G., et al. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc. Natl. Acad. Sci. USA. 2009;106:21848–21853. doi: 10.1073/pnas.0911354106. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

