Potent Inhibition of Human Cytomegalovirus by Modulation of Cellular SNARE Syntaxin 5
- PMID: 27795424
- PMCID: PMC5165218
- DOI: 10.1128/JVI.01637-16
Potent Inhibition of Human Cytomegalovirus by Modulation of Cellular SNARE Syntaxin 5
Abstract
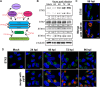
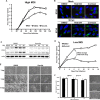
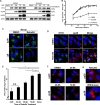
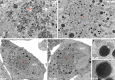
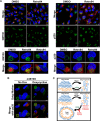
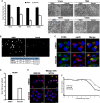
Formation of the cytoplasmic viral assembly compartment (cVAC) is an important step for efficient human cytomegalovirus (HCMV) assembly. To do this, the virus must alter and repurpose the normal cellular balance of membrane and protein flux, a process that is not well understood. Although a recent screen identified three viral proteins essential for cVAC formation, less is known about the contribution of cellular factors. We show that HCMV infection increases the protein level of a cellular trafficking factor, syntaxin 5 (STX5), a member of the syntaxin family of SNARE proteins. STX5 is recruited to the cVAC in infected cells and is required for the efficient production of infectious virions. We find that STX5 is important for normal cVAC morphology and the proper localization of viral proteins. A previously identified inhibitor of trafficking, Retro94, causes the mislocalization of STX5, an altered cVAC morphology, and dispersal of viral proteins. The presence of Retro94 results in severely impaired production of infectious virions, with a decrease as great as 5 logs. We show that this inhibition is conserved among different strains of HCMV and the various cell types that support infection, as well as for murine CMV. Thus, our data identify a key cellular trafficking factor important for supporting HCMV infection.
Importance: Human cytomegalovirus (HCMV) infection causes severe disease and mortality in immunocompromised individuals, including organ transplant and AIDS patients. In addition, infection of a developing fetus may result in lifelong complications such as deafness and learning disabilities. Understanding in detail the processes involved in HCMV replication is important for developing novel treatments. One of these essential processes, assembly of infectious virions, takes places in the cytoplasmic viral assembly compartment. We identify a cellular protein, syntaxin 5, important for generating this compartment, and show that it is required for the efficient production of infectious virions. We also show that a small molecule that disrupts this protein also significantly reduces the amount of infectious virions that are generated. Thus, by pinpointing a cellular protein that is important in the replication cycle of HCMV, we identified a novel target that can be pursued for therapeutic intervention.
Keywords: Golgi apparatus; HCMV; SNARE; assembly; assembly compartment; cytomegalovirus; protein trafficking.
Copyright © 2016 American Society for Microbiology.
Figures

Similar articles
-
Identification of human cytomegalovirus genes important for biogenesis of the cytoplasmic virion assembly complex.J Virol. 2014 Aug;88(16):9086-99. doi: 10.1128/JVI.01141-14. Epub 2014 Jun 4. J Virol. 2014. PMID: 24899189 Free PMC article.
-
Phosphorylation of Golgi Peripheral Membrane Protein Grasp65 Is an Integral Step in the Formation of the Human Cytomegalovirus Cytoplasmic Assembly Compartment.mBio. 2016 Oct 4;7(5):e01554-16. doi: 10.1128/mBio.01554-16. mBio. 2016. PMID: 27703074 Free PMC article.
-
Loss of the Human Cytomegalovirus US16 Protein Abrogates Virus Entry into Endothelial and Epithelial Cells by Reducing the Virion Content of the Pentamer.J Virol. 2017 May 12;91(11):e00205-17. doi: 10.1128/JVI.00205-17. Print 2017 Jun 1. J Virol. 2017. PMID: 28331097 Free PMC article.
-
Host Cell Signatures of the Envelopment Site within Beta-Herpes Virions.Int J Mol Sci. 2022 Sep 1;23(17):9994. doi: 10.3390/ijms23179994. Int J Mol Sci. 2022. PMID: 36077391 Free PMC article. Review.
-
Human Cytomegalovirus Egress: Overcoming Barriers and Co-Opting Cellular Functions.Viruses. 2021 Dec 22;14(1):15. doi: 10.3390/v14010015. Viruses. 2021. PMID: 35062219 Free PMC article. Review.
Cited by
-
Inhibition of diverse opportunistic viruses by structurally optimized retrograde trafficking inhibitors.Bioorg Med Chem. 2019 May 1;27(9):1795-1803. doi: 10.1016/j.bmc.2019.03.026. Epub 2019 Mar 13. Bioorg Med Chem. 2019. PMID: 30890396 Free PMC article.
-
Human Cytomegalovirus Envelope Protein gpUL132 Regulates Infectious Virus Production through Formation of the Viral Assembly Compartment.mBio. 2020 Sep 29;11(5):e02044-20. doi: 10.1128/mBio.02044-20. mBio. 2020. PMID: 32994323 Free PMC article.
-
Localization of the WD repeat-containing protein 5 to the Virion Assembly Compartment Facilitates Human Cytomegalovirus Assembly.J Virol. 2021 Mar 25;95(8):e02101-20. doi: 10.1128/JVI.02101-20. Epub 2021 Jan 27. J Virol. 2021. PMID: 33504601 Free PMC article.
-
Infection-Induced Changes Within the Endocytic Recycling Compartment Suggest a Roadmap of Human Cytomegalovirus Egress.Front Microbiol. 2018 Aug 22;9:1888. doi: 10.3389/fmicb.2018.01888. eCollection 2018. Front Microbiol. 2018. PMID: 30186245 Free PMC article.
-
Nonenvelopment Role for the ESCRT-III Complex during Human Cytomegalovirus Infection.J Virol. 2018 May 29;92(12):e02096-17. doi: 10.1128/JVI.02096-17. Print 2018 Jun 15. J Virol. 2018. PMID: 29618648 Free PMC article.
References
-
- Sanchez V, Greis KD, Sztul E, Britt WJ. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J Virol 74:975–986. doi:10.1128/JVI.74.2.975-986.2000. - DOI - PMC - PubMed
-
- Hook LM, Grey F, Grabski R, Tirabassi R, Doyle T, Hancock M, Landais I, Jeng S, McWeeney S, Britt W, Nelson JA. 2014. Cytomegalovirus miRNAs target secretory pathway genes to facilitate formation of the virion assembly compartment and reduce cytokine secretion. Cell Host Microbe 15:363–373. doi:10.1016/j.chom.2014.02.004. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

