Single-cell lineage tracing in the mammary gland reveals stochastic clonal dispersion of stem/progenitor cell progeny
- PMID: 27779190
- PMCID: PMC5093309
- DOI: 10.1038/ncomms13053
Single-cell lineage tracing in the mammary gland reveals stochastic clonal dispersion of stem/progenitor cell progeny
Abstract
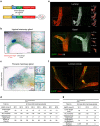
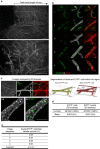
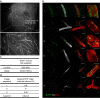
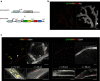
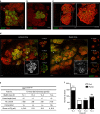
The mammary gland undergoes cycles of growth and regeneration throughout reproductive life, a process that requires mammary stem cells (MaSCs). Whilst recent genetic fate-mapping studies using lineage-specific promoters have provided valuable insights into the mammary epithelial hierarchy, the true differentiation potential of adult MaSCs remains unclear. To address this, herein we utilize a stochastic genetic-labelling strategy to indelibly mark a single cell and its progeny in situ, combined with tissue clearing and 3D imaging. Using this approach, clones arising from a single parent cell could be visualized in their entirety. We reveal that clonal progeny contribute exclusively to either luminal or basal lineages and are distributed sporadically to branching ducts or alveoli. Quantitative analyses suggest that pools of unipotent stem/progenitor cells contribute to adult mammary gland development. Our results highlight the utility of tracing a single cell and reveal that progeny of a single proliferative MaSC/progenitor are dispersed throughout the epithelium.
Figures

Similar articles
-
Techniques for the Reprogramming of Exogenous Stem/Progenitor Cell Populations Towards a Mammary Epithelial Cell Fate.Methods Mol Biol. 2017;1501:277-289. doi: 10.1007/978-1-4939-6475-8_14. Methods Mol Biol. 2017. PMID: 27796959 Free PMC article.
-
Neutral lineage tracing of proliferative embryonic and adult mammary stem/progenitor cells.Development. 2018 Jul 25;145(14):dev164079. doi: 10.1242/dev.164079. Development. 2018. PMID: 30045917 Free PMC article.
-
Murine mammary epithelial stem cells: discovery, function, and current status.Cold Spring Harb Perspect Biol. 2011 Feb 1;3(2):a004879. doi: 10.1101/cshperspect.a004879. Cold Spring Harb Perspect Biol. 2011. PMID: 20926515 Free PMC article. Review.
-
Developmental stage-specific contribution of LGR5(+) cells to basal and luminal epithelial lineages in the postnatal mammary gland.J Pathol. 2012 Nov;228(3):300-9. doi: 10.1002/path.4096. J Pathol. 2012. PMID: 22926799
-
The mammary stem cell hierarchy.Curr Top Dev Biol. 2014;107:133-60. doi: 10.1016/B978-0-12-416022-4.00005-6. Curr Top Dev Biol. 2014. PMID: 24439805 Review.
Cited by
-
Statistical theory of branching morphogenesis.Dev Growth Differ. 2018 Dec;60(9):512-521. doi: 10.1111/dgd.12570. Epub 2018 Oct 24. Dev Growth Differ. 2018. PMID: 30357803 Free PMC article. Review.
-
Fourteenth Annual ENBDC Workshop: Methods in Mammary Gland Biology and Breast Cancer.J Mammary Gland Biol Neoplasia. 2023 Oct 6;28(1):22. doi: 10.1007/s10911-023-09549-7. J Mammary Gland Biol Neoplasia. 2023. PMID: 37801168 Free PMC article.
-
Multidimensional Imaging of Mammary Gland Development: A Window Into Breast Form and Function.Front Cell Dev Biol. 2020 Mar 31;8:203. doi: 10.3389/fcell.2020.00203. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32296702 Free PMC article. Review.
-
Differentiation dynamics of mammary epithelial cells revealed by single-cell RNA sequencing.Nat Commun. 2017 Dec 11;8(1):2128. doi: 10.1038/s41467-017-02001-5. Nat Commun. 2017. PMID: 29225342 Free PMC article.
-
Long-lived unipotent Blimp1-positive luminal stem cells drive mammary gland organogenesis throughout adult life.Nat Commun. 2017 Nov 20;8(1):1714. doi: 10.1038/s41467-017-01971-w. Nat Commun. 2017. PMID: 29158490 Free PMC article.
References
-
- Watson C. J. & Khaled W. T. Mammary development in the embryo and adult: a journey of morphogenesis and commitment. Development 135, 995–1003 (2008). - PubMed
-
- Shackleton M. et al. Generation of a functional mammary gland from a single stem cell. Nature 439, 84–88 (2006). - PubMed
-
- Stingl J. et al. Purification and unique properties of mammary epithelial stem cells. Nature 439, 993–997 (2006). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

