Correcting mitochondrial fusion by manipulating mitofusin conformations
- PMID: 27775718
- PMCID: PMC5315023
- DOI: 10.1038/nature20156
Correcting mitochondrial fusion by manipulating mitofusin conformations
Abstract
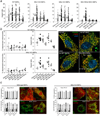
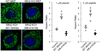
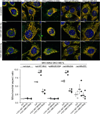
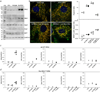
Mitochondria are dynamic organelles that exchange contents and undergo remodelling during cyclic fusion and fission. Genetic mutations in MFN2 (the gene encoding mitofusin 2) interrupt mitochondrial fusion and cause the untreatable neurodegenerative condition Charcot-Marie-Tooth disease type 2A (CMT2A). It has not yet been possible to directly modulate mitochondrial fusion, in part because the structural basis of mitofusin function is not completely understood. Here we show that mitofusins adopt either a fusion-constrained or a fusion-permissive molecular conformation, directed by specific intramolecular binding interactions, and demonstrate that mitofusin-dependent mitochondrial fusion can be regulated in mouse cells by targeting these conformational transitions. On the basis of this model, we engineered a cell-permeant minipeptide to destabilize the fusion-constrained conformation of mitofusin and promote the fusion-permissive conformation, reversing mitochondrial abnormalities in cultured fibroblasts and neurons that harbour CMT2A-associated genetic defects. The relationship between the conformational plasticity of mitofusin 2 and mitochondrial dynamism reveals a central mechanism that regulates mitochondrial fusion, the manipulation of which can correct mitochondrial pathology triggered by defective or imbalanced mitochondrial dynamics.
Figures




Similar articles
-
Mitochondrial Dysfunction and Pharmacodynamics of Mitofusin Activation in Murine Charcot-Marie-Tooth Disease Type 2A.J Pharmacol Exp Ther. 2022 Nov;383(2):137-148. doi: 10.1124/jpet.122.001332. Epub 2022 Sep 2. J Pharmacol Exp Ther. 2022. PMID: 36507849 Free PMC article.
-
A human mitofusin 2 mutation can cause mitophagic cardiomyopathy.Elife. 2023 Nov 1;12:e84235. doi: 10.7554/eLife.84235. Elife. 2023. PMID: 37910431 Free PMC article.
-
Burst mitofusin activation reverses neuromuscular dysfunction in murine CMT2A.Elife. 2020 Oct 19;9:e61119. doi: 10.7554/eLife.61119. Elife. 2020. PMID: 33074106 Free PMC article.
-
Charcot-Marie-Tooth type 2A in vivo models: Current updates.J Cell Mol Med. 2024 May;28(9):e18293. doi: 10.1111/jcmm.18293. J Cell Mol Med. 2024. PMID: 38722298 Free PMC article. Review.
-
MFN2-related neuropathies: Clinical features, molecular pathogenesis and therapeutic perspectives.J Neurol Sci. 2015 Sep 15;356(1-2):7-18. doi: 10.1016/j.jns.2015.05.033. Epub 2015 May 29. J Neurol Sci. 2015. PMID: 26143526 Review.
Cited by
-
Chemical Modulation of Mitochondria-Endoplasmic Reticulum Contact Sites.Cells. 2020 Jul 7;9(7):1637. doi: 10.3390/cells9071637. Cells. 2020. PMID: 32646031 Free PMC article. Review.
-
Acquired Expression of Mutant Mitofusin 2 Causes Progressive Neurodegeneration and Abnormal Behavior.J Neurosci. 2019 Feb 27;39(9):1588-1604. doi: 10.1523/JNEUROSCI.2139-18.2018. Epub 2019 Jan 3. J Neurosci. 2019. PMID: 30606759 Free PMC article.
-
Mitochondrial division, fusion and degradation.J Biochem. 2020 Mar 1;167(3):233-241. doi: 10.1093/jb/mvz106. J Biochem. 2020. PMID: 31800050 Free PMC article. Review.
-
Cardiac-specific research platforms engender novel insights into mitochondrial dynamism.Curr Opin Physiol. 2018 Jun;3:110-115. doi: 10.1016/j.cophys.2018.03.006. Epub 2018 Mar 26. Curr Opin Physiol. 2018. PMID: 30467553 Free PMC article.
-
Proteolytic regulation of mitochondrial dynamics.Mitochondrion. 2019 Nov;49:289-304. doi: 10.1016/j.mito.2019.04.008. Epub 2019 Apr 25. Mitochondrion. 2019. PMID: 31029640 Free PMC article. Review.
References
-
- Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46:265–287. - PubMed
-
- Kasahara A, Cipolat S, Chen Y, Dorn GW, 2nd, Scorrano L. Mitochondrial fusion directs cardiomyocyte differentiation via calcineurin and Notch signaling. Science. 2013;342:734–737. - PubMed
-
- Chen H, McCaffery JM, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–562. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

