Human cytomegalovirus reactivation from latency: validation of a "switch" model in vitro
- PMID: 27770817
- PMCID: PMC5075216
- DOI: 10.1186/s12985-016-0634-z
Human cytomegalovirus reactivation from latency: validation of a "switch" model in vitro
Abstract
Background: Human cytomegalovirus (HCMV) is an opportunistic pathogen leading to severe and even fatal diseases in 'at-risk' categories of individuals upon primary infection or the symptomatic reactivation of the endogenous virus. The mechanisms which make the virus able to reactivate from latency are still matter of intense study. However, the very low number of peripheral blood monocytes (an important latent virus reservoir) harbouring HCMV DNA makes it very difficult to obtain adequate viral quantities to use in such studies. Thus, the aim of the present study was to demonstrate the usefulness of human THP-1 monocytes, mostly employed as HCMV latent or lytic infection system, as a reactivation model.
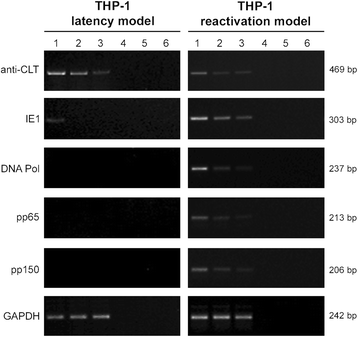
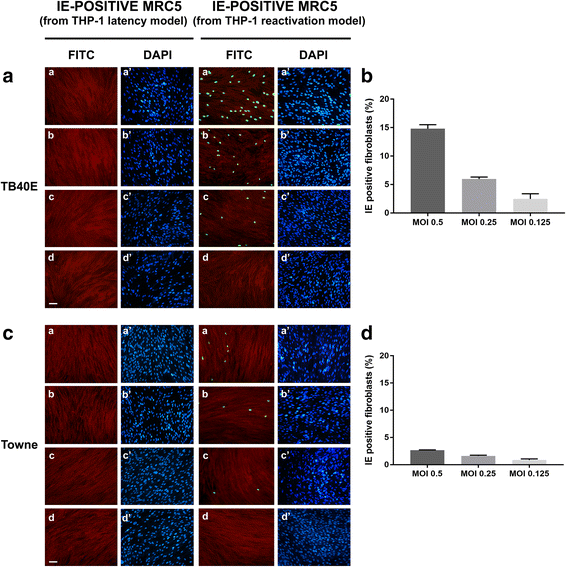
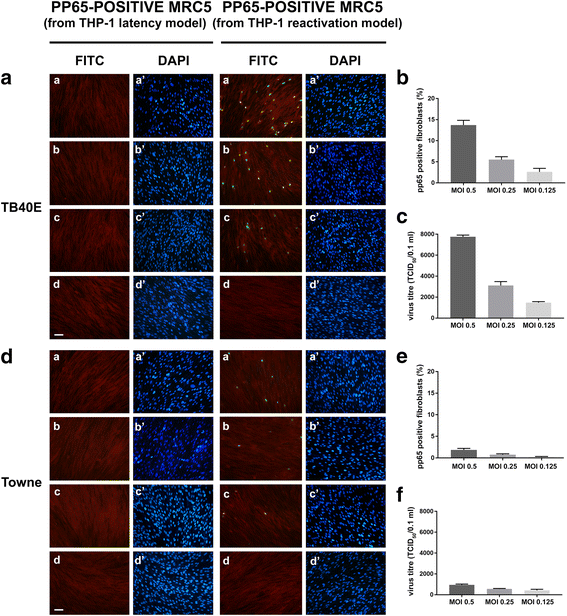
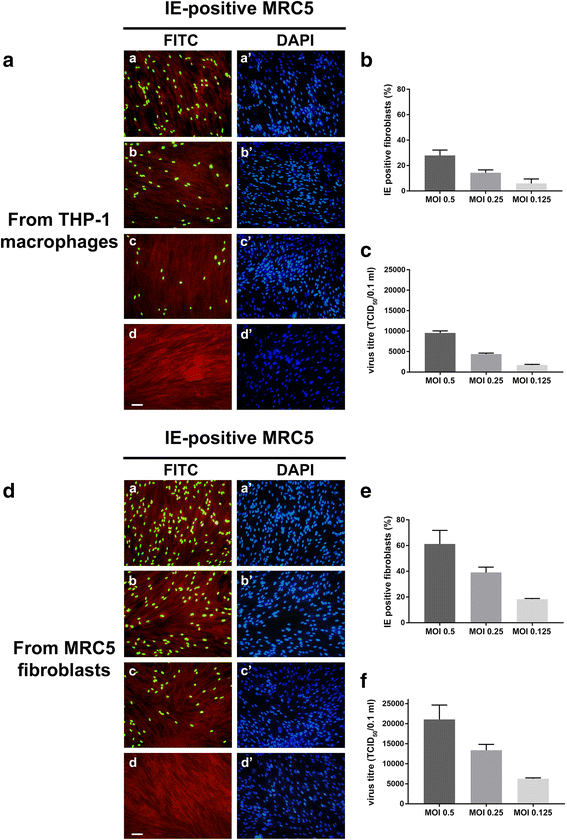
Methods: THP-1 monocytes were infected with HCMV TB40E strain (latency model) at multiplicities of infection (MOI) of 0.5, 0.25 or 0.125. After infection, THP-1 aliquots were differentiated into macrophages (reactivation model). Infections were carried out for 30 h, 4, 6 and 7 days. Viral DNA evaluation was performed with viable and UV-inactivated virus by q-Real-Time PCR. RNA extracted from latency and reactivation models at 7 days post-infection (p.i.) was subjected to RT-PCR to analyse viral latency and lytic transcripts. To perform viral progeny analysis and titration, the culture medium from infected THP-1 latency and reactivation models (7 days p.i.) was used to infect human fibroblasts; it was also checked for the presence of exosomes. For viral progeny analysis experiments, the Towne strain was also used.
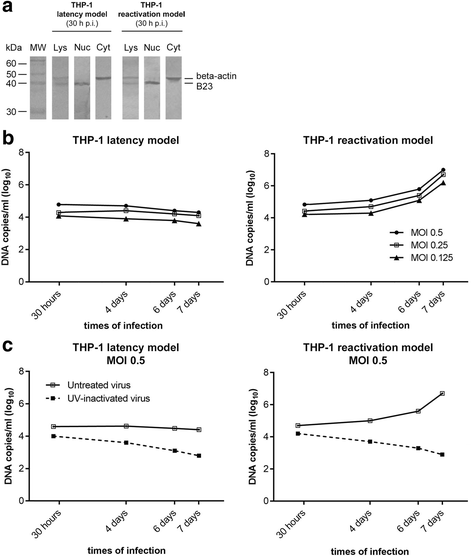
Results: Our results showed that, while comparable TB40E DNA amounts were present in both latent and reactivation models at 30 h p.i., gradually increased quantities of viral DNA were only evident in the latter model at 4, 6, 7 days p.i.. The completion of the lytic cycle upon reactivation was also proved by the presence of HCMV lytic transcripts and an infectious viral yield at 7 days p.i.
Conclusions: Our data demonstrate the effectiveness of THP-1 cells as a "switch" model for studying the mechanisms that regulate HCMV reactivation from latency. This system is able to provide adequate quantities of cells harbouring latent/reactivated virus, thereby overcoming the intrinsic difficulties connected to the ex vivo system.
Keywords: HCMV; Latency; Reactivation; THP-1 differentiation; THP-1 monocytes.
Figures
Similar articles
-
Latency-Associated Expression of Human Cytomegalovirus US28 Attenuates Cell Signaling Pathways To Maintain Latent Infection.mBio. 2017 Dec 5;8(6):e01754-17. doi: 10.1128/mBio.01754-17. mBio. 2017. PMID: 29208743 Free PMC article.
-
Human Cytomegalovirus US28 Ligand Binding Activity Is Required for Latency in CD34+ Hematopoietic Progenitor Cells and Humanized NSG Mice.mBio. 2019 Aug 20;10(4):e01889-19. doi: 10.1128/mBio.01889-19. mBio. 2019. PMID: 31431555 Free PMC article.
-
Human cytomegalovirus modulates monocyte-mediated innate immune responses during short-term experimental latency in vitro.J Virol. 2014 Aug;88(16):9391-405. doi: 10.1128/JVI.00934-14. Epub 2014 Jun 11. J Virol. 2014. PMID: 24920803 Free PMC article.
-
Aspects of human cytomegalovirus latency and reactivation.Curr Top Microbiol Immunol. 2008;325:297-313. doi: 10.1007/978-3-540-77349-8_17. Curr Top Microbiol Immunol. 2008. PMID: 18637513 Review.
-
Chromatin structure regulates human cytomegalovirus gene expression during latency, reactivation and lytic infection.Biochim Biophys Acta. 2010 Mar-Apr;1799(3-4):286-95. doi: 10.1016/j.bbagrm.2009.08.001. Epub 2009 Aug 12. Biochim Biophys Acta. 2010. PMID: 19682613 Review.
Cited by
-
CD34+ Hematopoietic Progenitor Cell Subsets Exhibit Differential Ability To Maintain Human Cytomegalovirus Latency and Persistence.J Virol. 2021 Jan 13;95(3):e02105-20. doi: 10.1128/JVI.02105-20. Print 2021 Jan 13. J Virol. 2021. PMID: 33177198 Free PMC article.
-
Different expression pattern of human cytomegalovirus-encoded microRNAs in circulation from virus latency to reactivation.J Transl Med. 2020 Dec 9;18(1):469. doi: 10.1186/s12967-020-02653-w. J Transl Med. 2020. PMID: 33298092 Free PMC article.
-
Cytomegalovirus-induced peroxynitrite promotes virus entry and contributes to pathogenesis in a murine model of infection.mBio. 2024 Aug 14;15(8):e0315223. doi: 10.1128/mbio.03152-23. Epub 2024 Jul 2. mBio. 2024. PMID: 38953361 Free PMC article.
-
Systems Virology and Human Cytomegalovirus: Using High Throughput Approaches to Identify Novel Host-Virus Interactions During Lytic Infection.Front Cell Infect Microbiol. 2020 Jun 10;10:280. doi: 10.3389/fcimb.2020.00280. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32587832 Free PMC article. Review.
-
A Virally Encoded DeSUMOylase Activity Is Required for Cytomegalovirus Reactivation from Latency.Cell Rep. 2018 Jul 17;24(3):594-606. doi: 10.1016/j.celrep.2018.06.048. Cell Rep. 2018. PMID: 30021158 Free PMC article.
References
-
- Mocarski ES, Tan Courcelle C. Cytomegaloviruses and their replication. In: Howley DMKPM, editor. Fields Virology. 4. Philadelphia: Lippincott Williams and Wilkins; 2001. pp. 2629–2673.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

