Crystal Structure of the Human Cannabinoid Receptor CB1
- PMID: 27768894
- PMCID: PMC5322940
- DOI: 10.1016/j.cell.2016.10.004
Crystal Structure of the Human Cannabinoid Receptor CB1
Abstract
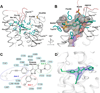
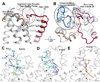
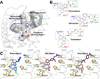
Cannabinoid receptor 1 (CB1) is the principal target of Δ9-tetrahydrocannabinol (THC), a psychoactive chemical from Cannabis sativa with a wide range of therapeutic applications and a long history of recreational use. CB1 is activated by endocannabinoids and is a promising therapeutic target for pain management, inflammation, obesity, and substance abuse disorders. Here, we present the 2.8 Å crystal structure of human CB1 in complex with AM6538, a stabilizing antagonist, synthesized and characterized for this structural study. The structure of the CB1-AM6538 complex reveals key features of the receptor and critical interactions for antagonist binding. In combination with functional studies and molecular modeling, the structure provides insight into the binding mode of naturally occurring CB1 ligands, such as THC, and synthetic cannabinoids. This enhances our understanding of the molecular basis for the physiological functions of CB1 and provides new opportunities for the design of next-generation CB1-targeting pharmaceuticals.
Keywords: AM6538; G protein-coupled receptor; THC; cannabinoid receptor CB1; cell signalling; crystal structure; marijuana; rimonabant; stabilizing antagonist.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
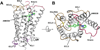

Comment in
-
Drug design: Cannabinoid receptor structure revealed.Nat Rev Drug Discov. 2016 Dec;15(12):822. doi: 10.1038/nrd.2016.242. Epub 2016 Nov 18. Nat Rev Drug Discov. 2016. PMID: 27857139 No abstract available.
Similar articles
-
Probing the CB1 Cannabinoid Receptor Binding Pocket with AM6538, a High-Affinity Irreversible Antagonist.Mol Pharmacol. 2019 Nov;96(5):619-628. doi: 10.1124/mol.119.116483. Epub 2019 Sep 12. Mol Pharmacol. 2019. PMID: 31515283 Free PMC article.
-
Crystal structures of agonist-bound human cannabinoid receptor CB1.Nature. 2017 Jul 27;547(7664):468-471. doi: 10.1038/nature23272. Epub 2017 Jul 5. Nature. 2017. PMID: 28678776 Free PMC article.
-
Ligand-specific homology modeling of human cannabinoid (CB1) receptor.J Mol Graph Model. 2012 Sep;38:155-64. doi: 10.1016/j.jmgm.2012.05.002. Epub 2012 May 31. J Mol Graph Model. 2012. PMID: 23079645
-
[Drug discrimination properties and cytotoxicity of the cannabinoid receptor ligands].Nihon Arukoru Yakubutsu Igakkai Zasshi. 2012 Jun;47(3):135-43. Nihon Arukoru Yakubutsu Igakkai Zasshi. 2012. PMID: 22894054 Review. Japanese.
-
Cannabinoid CB1 and CB2 receptor ligand specificity and the development of CB2-selective agonists.Curr Med Chem. 2008;15(14):1428-43. doi: 10.2174/092986708784567716. Curr Med Chem. 2008. PMID: 18537620 Review.
Cited by
-
Evaluating Allosteric Perturbations in Cannabinoid Receptor 1 by In Silico Single-Point Mutation.ACS Omega. 2022 Oct 14;7(42):37873-37884. doi: 10.1021/acsomega.2c04980. eCollection 2022 Oct 25. ACS Omega. 2022. PMID: 36312415 Free PMC article.
-
Perspectives on Challenges in Cannabis Drug Delivery Systems: Where Are We?Med Cannabis Cannabinoids. 2022 Jul 25;5(1):102-119. doi: 10.1159/000525629. eCollection 2022 Jan-Dec. Med Cannabis Cannabinoids. 2022. PMID: 36467783 Free PMC article. Review.
-
N-[1,3-Dialkyl(aryl)-2-oxoimidazolidin-4-ylidene]-aryl(alkyl)sulphonamides as Novel Selective Human Cannabinoid Type 2 Receptor (hCB2R) Ligands; Insights into the Mechanism of Receptor Activation/Deactivation.Molecules. 2022 Nov 23;27(23):8152. doi: 10.3390/molecules27238152. Molecules. 2022. PMID: 36500256 Free PMC article.
-
Structures of the Human PGD2 Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition.Mol Cell. 2018 Oct 4;72(1):48-59.e4. doi: 10.1016/j.molcel.2018.08.009. Epub 2018 Sep 13. Mol Cell. 2018. PMID: 30220562 Free PMC article.
-
Cannabinoids in Medicine: A Multifaceted Exploration of Types, Therapeutic Applications, and Emerging Opportunities in Neurodegenerative Diseases and Cancer Therapy.Biomolecules. 2023 Sep 14;13(9):1388. doi: 10.3390/biom13091388. Biomolecules. 2023. PMID: 37759788 Free PMC article. Review.
References
-
- Abagyan R, Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J Mol Biol. 1994;235:983–1002. - PubMed
-
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25.
-
- Adams R, Mac KS, Jr, Loewe S. Tetrahydrocannabinol homologs with double branched alkyl groups in the 3-position. J Am Chem Soc. 1948;70:664–668. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

