Reproducibility of Digital PCR Assays for Circulating Tumor DNA Analysis in Advanced Breast Cancer
- PMID: 27760227
- PMCID: PMC5070760
- DOI: 10.1371/journal.pone.0165023
Reproducibility of Digital PCR Assays for Circulating Tumor DNA Analysis in Advanced Breast Cancer
Abstract
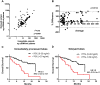
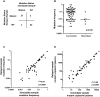
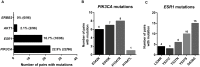
Circulating tumor DNA (ctDNA) analysis has the potential to allow non-invasive analysis of tumor mutations in advanced cancer. In this study we assessed the reproducibility of digital PCR (dPCR) assays of circulating tumor DNA in a cohort of patients with advanced breast cancer and assessed delayed plasma processing using cell free DNA preservative tubes. We recruited a cohort of 96 paired samples from 71 women with advanced breast cancer who had paired blood samples processed either immediately or delayed in preservative tubes with processing 48-72 hours after collection. Plasma DNA was analysed with multiplex digital PCR (mdPCR) assays for hotspot mutations in PIK3CA, ESR1 and ERBB2, and for AKT1 E17K. There was 94.8% (91/96) agreement in mutation calling between immediate and delayed processed tubes, kappa 0.88 95% CI 0.77-0.98). Discordance in mutation calling resulted from low allele frequency and likely stochastic effects. In concordant samples there was high correlation in mutant copies per ml plasma (r2 = 0.98; p<0.0001). There was elevation of total cell free plasma DNA concentrations in 10.3% of delayed processed tubes, although overall quantification of total cell free plasma DNA had similar prognostic effects in immediate (HR 3.6) and delayed (HR 3.0) tubes. There was moderate agreement in changes in allele fraction between sequential samples in quantitative mutation tracking (r = 0.84, p = 0.0002). Delayed processing of samples using preservative tubes allows for centralized ctDNA digital PCR mutation screening in advanced breast cancer. The potential of preservative tubes in quantitative mutation tracking requires further research.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Comparison of cell stabilizing blood collection tubes for circulating plasma tumor DNA.Clin Biochem. 2015 Oct;48(15):993-8. doi: 10.1016/j.clinbiochem.2015.07.097. Epub 2015 Jul 31. Clin Biochem. 2015. PMID: 26234639 Free PMC article.
-
Tumor burden monitoring using cell-free tumor DNA could be limited by tumor heterogeneity in advanced breast cancer and should be evaluated together with radiographic imaging.BMC Cancer. 2017 Mar 22;17(1):210. doi: 10.1186/s12885-017-3185-9. BMC Cancer. 2017. PMID: 28330468 Free PMC article.
-
Comparison of BEAMing and Droplet Digital PCR for Circulating Tumor DNA Analysis.Clin Chem. 2019 Nov;65(11):1405-1413. doi: 10.1373/clinchem.2019.305805. Epub 2019 Sep 24. Clin Chem. 2019. PMID: 31551314
-
Use of cell free DNA in breast oncology.Biochim Biophys Acta. 2016 Apr;1865(2):266-74. doi: 10.1016/j.bbcan.2016.03.006. Epub 2016 Mar 22. Biochim Biophys Acta. 2016. PMID: 27012505 Review.
-
Circulating Plasma Tumor DNA.Adv Exp Med Biol. 2016;882:259-76. doi: 10.1007/978-3-319-22909-6_11. Adv Exp Med Biol. 2016. PMID: 26987539 Review.
Cited by
-
The Impact of ESR1 Mutations on the Treatment of Metastatic Breast Cancer.Horm Cancer. 2018 Aug;9(4):215-228. doi: 10.1007/s12672-017-0306-5. Epub 2018 May 7. Horm Cancer. 2018. PMID: 29736566 Free PMC article. Review.
-
From Sampling to Sequencing: A Liquid Biopsy Pre-Analytic Workflow to Maximize Multi-Layer Genomic Information from a Single Tube.Cancers (Basel). 2021 Jun 15;13(12):3002. doi: 10.3390/cancers13123002. Cancers (Basel). 2021. PMID: 34203921 Free PMC article.
-
Pre-Analytical Evaluation of Streck Cell-Free DNA Blood Collection Tubes for Liquid Profiling in Oncology.Diagnostics (Basel). 2023 Mar 29;13(7):1288. doi: 10.3390/diagnostics13071288. Diagnostics (Basel). 2023. PMID: 37046506 Free PMC article.
-
Specialized Blood Collection Tubes for Liquid Biopsy: Improving the Pre-analytical Conditions.Mol Diagn Ther. 2020 Feb;24(1):113-124. doi: 10.1007/s40291-019-00442-w. Mol Diagn Ther. 2020. PMID: 31838654
-
Impact of Molecular Profiling on Therapy Management in Breast Cancer.J Clin Med. 2024 Aug 23;13(17):4995. doi: 10.3390/jcm13174995. J Clin Med. 2024. PMID: 39274207 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

