Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy
- PMID: 27760051
- PMCID: PMC5096930
- DOI: 10.1172/JCI87927
Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy
Abstract
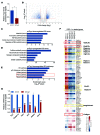
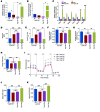
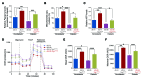
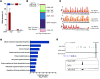
The regulatory roles of long noncoding RNAs (lncRNAs) in transcriptional coactivators are still largely unknown. Here, we have shown that the peroxisome proliferator-activated receptor γ (PPARγ) coactivator α (PGC-1α, encoded by Ppargc1a) is functionally regulated by the lncRNA taurine-upregulated gene 1 (Tug1). Further, we have described a role for Tug1 in the regulation of mitochondrial function in podocytes. Using a murine model of diabetic nephropathy (DN), we performed an unbiased RNA-sequencing (RNA-seq) analysis of kidney glomeruli and identified Tug1 as a differentially expressed lncRNA in the diabetic milieu. Podocyte-specific overexpression (OE) of Tug1 in diabetic mice improved the biochemical and histological features associated with DN. Unexpectedly, we found that Tug1 OE rescued the expression of PGC-1α and its transcriptional targets. Tug1 OE was also associated with improvements in mitochondrial bioenergetics in the podocytes of diabetic mice. Mechanistically, we found that the interaction between Tug1 and PGC-1α promotes the binding of PGC-1α to its own promoter. We identified a Tug1-binding element (TBE) upstream of the Ppargc1a gene and showed that Tug1 binds with the TBE to enhance Ppargc1a promoter activity. These findings indicate that a direct interaction between PGC-1α and Tug1 modulates mitochondrial bioenergetics in podocytes in the diabetic milieu.
Figures


Comment in
-
The long noncoding RNA Tug1 connects metabolic changes with kidney disease in podocytes.J Clin Invest. 2016 Nov 1;126(11):4072-4075. doi: 10.1172/JCI90828. Epub 2016 Oct 17. J Clin Invest. 2016. PMID: 27760046 Free PMC article.
Similar articles
-
The long noncoding RNA Tug1 connects metabolic changes with kidney disease in podocytes.J Clin Invest. 2016 Nov 1;126(11):4072-4075. doi: 10.1172/JCI90828. Epub 2016 Oct 17. J Clin Invest. 2016. PMID: 27760046 Free PMC article.
-
PGC1α is required for the renoprotective effect of lncRNA Tug1 in vivo and links Tug1 with urea cycle metabolites.Cell Rep. 2021 Aug 10;36(6):109510. doi: 10.1016/j.celrep.2021.109510. Cell Rep. 2021. PMID: 34380028 Free PMC article.
-
Long non-coding RNA Tug1 modulates mitochondrial and myogenic responses to exercise in skeletal muscle.BMC Biol. 2022 Jul 18;20(1):164. doi: 10.1186/s12915-022-01366-4. BMC Biol. 2022. PMID: 35850762 Free PMC article.
-
The Role of Peroxisome Proliferator-Activated Receptor γ Coactivator 1α (PGC-1α) in Kidney Disease.Semin Nephrol. 2018 Mar;38(2):121-126. doi: 10.1016/j.semnephrol.2018.01.003. Semin Nephrol. 2018. PMID: 29602395 Free PMC article. Review.
-
Podocyte Bioenergetics in the Development of Diabetic Nephropathy: The Role of Mitochondria.Endocrinology. 2022 Jan 1;163(1):bqab234. doi: 10.1210/endocr/bqab234. Endocrinology. 2022. PMID: 34791124 Free PMC article. Review.
Cited by
-
M6A-mediated upregulation of lncRNA TUG1 in liver cancer cells regulates the antitumor response of CD8+ T cells and phagocytosis of macrophages.Adv Sci (Weinh). 2024 Sep;11(34):e2400695. doi: 10.1002/advs.202400695. Epub 2024 Jul 9. Adv Sci (Weinh). 2024. PMID: 38981064 Free PMC article.
-
lncRNA TUG1 regulates hyperuricemia-induced renal fibrosis in a rat model.Acta Biochim Biophys Sin (Shanghai). 2022 Sep 25;54(9):1365-1375. doi: 10.3724/abbs.2022128. Acta Biochim Biophys Sin (Shanghai). 2022. PMID: 36148952 Free PMC article.
-
LncTUG1 ameliorates renal tubular fibrosis in experimental diabetic nephropathy through the miR-145-5p/dual-specificity phosphatase 6 axis.Ren Fail. 2023 Dec;45(1):2173950. doi: 10.1080/0886022X.2023.2173950. Ren Fail. 2023. PMID: 36794657 Free PMC article.
-
Dysregulated expression but redundant function of the long non-coding RNA HOTAIR in diabetic kidney disease.Diabetologia. 2019 Nov;62(11):2129-2142. doi: 10.1007/s00125-019-4967-1. Epub 2019 Aug 9. Diabetologia. 2019. PMID: 31399844
-
LncRNA GACAT2 binds with protein PKM1/2 to regulate cell mitochondrial function and cementogenesis in an inflammatory environment.Bone Res. 2022 Mar 16;10(1):29. doi: 10.1038/s41413-022-00197-x. Bone Res. 2022. PMID: 35296649 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

