Understanding cardiomyocyte proliferation: an insight into cell cycle activity
- PMID: 27695872
- PMCID: PMC11107761
- DOI: 10.1007/s00018-016-2375-y
Understanding cardiomyocyte proliferation: an insight into cell cycle activity
Abstract
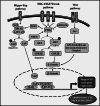
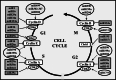
Cardiomyocyte proliferation and regeneration are key to the functional recovery of myocardial tissue from injury. In the recent years, studies on cardiomyocyte proliferation overturned the traditional belief that adult cardiomyocytes permanently withdraw from the cell cycle activity. Hence, targeting cardiomyocyte proliferation is one of the potential therapeutic strategies for myocardial regeneration and repair. To achieve this, a deep understanding of the fundamental mechanisms involved in cardiomyocyte cell cycle as well as differences between neonatal and adult cardiomyocytes' cell cycle activity is required. This review focuses on the recent progress in understanding of cardiomyocyte cell cycle activity at different life stages viz., gestation, birth, and adulthood. The temporal expression/activities of major cell cycle activators (cyclins and CDKs), inhibitors (p21, p27, p57, p16, and p18), and cell-cycle-associated proteins (Rb, p107, and p130) in cardiomyocytes during gestation and postnatal life are described in this review. The influence of different transcription factors and microRNAs on the expression of cell cycle proteins is demonstrated. This review also deals major pathways (PI3K/AKT, Wnt/β-catenin, and Hippo-YAP) associated with cardiomyocyte cell cycle progression. Furthermore, the postnatal alterations in structure and cellular events responsible for the loss of cell cycle activity are also illustrated.
Keywords: Cardiomyocytes; Cell cycle; Cyclins; MicroRNAs; Signaling pathways; Transcription factors.
Figures
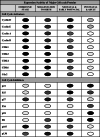
 indicates higher expression and activity. Lighter shades
indicates higher expression and activity. Lighter shades
 indicate comparatively decreased/lower activity. Empty circles
indicate comparatively decreased/lower activity. Empty circles
 indicate barely detectable or undetectable. CDK cyclin-dependent kinase, CDC cell division cycle protein, Rb retinoblastoma
indicate barely detectable or undetectable. CDK cyclin-dependent kinase, CDC cell division cycle protein, Rb retinoblastoma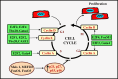
Similar articles
-
Melatonin promotes cardiomyocyte proliferation and heart repair in mice with myocardial infarction via miR-143-3p/Yap/Ctnnd1 signaling pathway.Acta Pharmacol Sin. 2021 Jun;42(6):921-931. doi: 10.1038/s41401-020-0495-2. Epub 2020 Aug 24. Acta Pharmacol Sin. 2021. PMID: 32839503 Free PMC article.
-
Defining the molecular underpinnings controlling cardiomyocyte proliferation.Clin Sci (Lond). 2022 Jun 30;136(12):911-934. doi: 10.1042/CS20211180. Clin Sci (Lond). 2022. PMID: 35723259 Review.
-
A microRNA-Hippo pathway that promotes cardiomyocyte proliferation and cardiac regeneration in mice.Sci Transl Med. 2015 Mar 18;7(279):279ra38. doi: 10.1126/scitranslmed.3010841. Sci Transl Med. 2015. PMID: 25787764 Free PMC article.
-
mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts.Circ Res. 2013 Jun 7;112(12):1557-66. doi: 10.1161/CIRCRESAHA.112.300658. Epub 2013 Apr 10. Circ Res. 2013. PMID: 23575307 Free PMC article.
-
Upstream regulation of the Hippo-Yap pathway in cardiomyocyte regeneration.Semin Cell Dev Biol. 2020 Apr;100:11-19. doi: 10.1016/j.semcdb.2019.09.004. Epub 2019 Oct 9. Semin Cell Dev Biol. 2020. PMID: 31606277 Free PMC article. Review.
Cited by
-
Meta-analysis of Transcriptomic Data Reveals Pathophysiological Modules Involved with Atrial Fibrillation.Mol Diagn Ther. 2020 Dec;24(6):737-751. doi: 10.1007/s40291-020-00497-0. Epub 2020 Oct 23. Mol Diagn Ther. 2020. PMID: 33095430
-
Wound Healing Potential of Spirulina Protein on CCD-986sk Cells.Mar Drugs. 2019 Feb 22;17(2):130. doi: 10.3390/md17020130. Mar Drugs. 2019. PMID: 30813318 Free PMC article.
-
Ploidy-stratified single cardiomyocyte transcriptomics map Zinc Finger E-Box Binding Homeobox 1 to underly cardiomyocyte proliferation before birth.Basic Res Cardiol. 2023 Mar 2;118(1):8. doi: 10.1007/s00395-023-00979-2. Basic Res Cardiol. 2023. PMID: 36862248 Free PMC article.
-
The Aurora Kinase Inhibitor CYC116 Promotes the Maturation of Cardiomyocytes Derived from Human Pluripotent Stem Cells.Mol Cells. 2022 Dec 31;45(12):923-934. doi: 10.14348/molcells.2022.0077. Epub 2022 Dec 13. Mol Cells. 2022. PMID: 36572561 Free PMC article.
-
TMEM11 regulates cardiomyocyte proliferation and cardiac repair via METTL1-mediated m7G methylation of ATF5 mRNA.Cell Death Differ. 2023 Jul;30(7):1786-1798. doi: 10.1038/s41418-023-01179-0. Epub 2023 Jun 7. Cell Death Differ. 2023. PMID: 37286744 Free PMC article.
References
-
- Erokhina IL, Selivanova GV, Vlasova TD, Emel’ianova OI. Correlation between the level of polyploidy and hypertrophy and degree of human atrial cardiomyocyte damage in certain congenital and acquired heart pathologies. Tsitologiia. 1997;39:889–899. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

