CRISPR/Cas9 Targets Chicken Embryonic Somatic Cells In Vitro and In Vivo and generates Phenotypic Abnormalities
- PMID: 27694906
- PMCID: PMC5046125
- DOI: 10.1038/srep34524
CRISPR/Cas9 Targets Chicken Embryonic Somatic Cells In Vitro and In Vivo and generates Phenotypic Abnormalities
Abstract
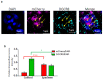
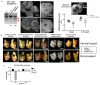
Chickens are an invaluable model for studying human diseases, physiology and especially development, but have lagged in genetic applications. With the advent of Programmable Engineered Nucleases, genetic manipulation has become efficient, specific and rapid. Here, we show that the CRISPR/Cas9 system can precisely edit the chicken genome. We generated HIRA, TYRP1, DICER, MBD3, EZH2, and 6 other gene knockouts in two chicken cell lines using the CRISPR/Cas9 system, with no off-target effects detected. We also showed that very large deletions (>75 kb) could be achieved. We also achieved targeted modification by homology-directed repair (HDR), producing MEN2A and MEN2B mutations of the RET gene. We also targeted DGCR8 in neural cells of the chicken embryo by in vivo electroporation. After FACS isolation of transfected cells, we observed appropriate sequence changes in DGCR8. Wholemount and frozen section antibody labelling showed reduction of DGCR8 levels in transfected cells. In addition, there was reduced expression levels of DGCR8-associated genes DROSHA, YPEL1 and NGN2. We also observed morphological differences in neural tissue and cardiac-related tissues of transfected embryos. These findings demonstrate that precisely targeted genetic manipulation of the genome using the CRISPR/Cas9 system can be extended to the highly adaptable in vivo chicken embryo model.
Figures


Similar articles
-
Targeted mutagenesis in chicken using CRISPR/Cas9 system.Sci Rep. 2016 Apr 6;6:23980. doi: 10.1038/srep23980. Sci Rep. 2016. PMID: 27050479 Free PMC article.
-
CRISPR mediated somatic cell genome engineering in the chicken.Dev Biol. 2015 Nov 1;407(1):68-74. doi: 10.1016/j.ydbio.2015.08.007. Epub 2015 Aug 13. Dev Biol. 2015. PMID: 26277216
-
Efficient edition of the bovine PRNP prion gene in somatic cells and IVF embryos using the CRISPR/Cas9 system.Theriogenology. 2016 Nov;86(8):1886-1896.e1. doi: 10.1016/j.theriogenology.2016.06.010. Epub 2016 Jun 15. Theriogenology. 2016. PMID: 27566851
-
CRISPR/Cas9-mediated correction of human genetic disease.Sci China Life Sci. 2017 May;60(5):447-457. doi: 10.1007/s11427-017-9032-4. Epub 2017 May 3. Sci China Life Sci. 2017. PMID: 28534256 Review.
-
The application of genome editing in studying hearing loss.Hear Res. 2015 Sep;327:102-8. doi: 10.1016/j.heares.2015.04.016. Epub 2015 May 15. Hear Res. 2015. PMID: 25987504 Free PMC article. Review.
Cited by
-
Genome editing of avian species: implications for animal use and welfare.Lab Anim. 2022 Feb;56(1):50-59. doi: 10.1177/0023677221998400. Epub 2021 Mar 10. Lab Anim. 2022. PMID: 33691522 Free PMC article.
-
Study of the regulatory elements of the Ovalbumin gene promoter using CRISPR technology in chicken cells.J Biol Eng. 2023 Jul 17;17(1):46. doi: 10.1186/s13036-023-00367-3. J Biol Eng. 2023. PMID: 37461059 Free PMC article.
-
Follow Me! A Tale of Avian Heart Development with Comparisons to Mammal Heart Development.J Cardiovasc Dev Dis. 2020 Mar 7;7(1):8. doi: 10.3390/jcdd7010008. J Cardiovasc Dev Dis. 2020. PMID: 32156044 Free PMC article. Review.
-
Gene editing in birds takes flight.Mamm Genome. 2017 Aug;28(7-8):315-323. doi: 10.1007/s00335-017-9701-z. Epub 2017 Jun 13. Mamm Genome. 2017. PMID: 28612238 Free PMC article. Review.
-
Systematic analysis of transcription start sites in avian development.PLoS Biol. 2017 Sep 5;15(9):e2002887. doi: 10.1371/journal.pbio.2002887. eCollection 2017 Sep. PLoS Biol. 2017. PMID: 28873399 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

