Age-related accrual of methylomic variability is linked to fundamental ageing mechanisms
- PMID: 27654999
- PMCID: PMC5032245
- DOI: 10.1186/s13059-016-1053-6
Age-related accrual of methylomic variability is linked to fundamental ageing mechanisms
Abstract
Background: Epigenetic change is a hallmark of ageing but its link to ageing mechanisms in humans remains poorly understood. While DNA methylation at many CpG sites closely tracks chronological age, DNA methylation changes relevant to biological age are expected to gradually dissociate from chronological age, mirroring the increased heterogeneity in health status at older ages.
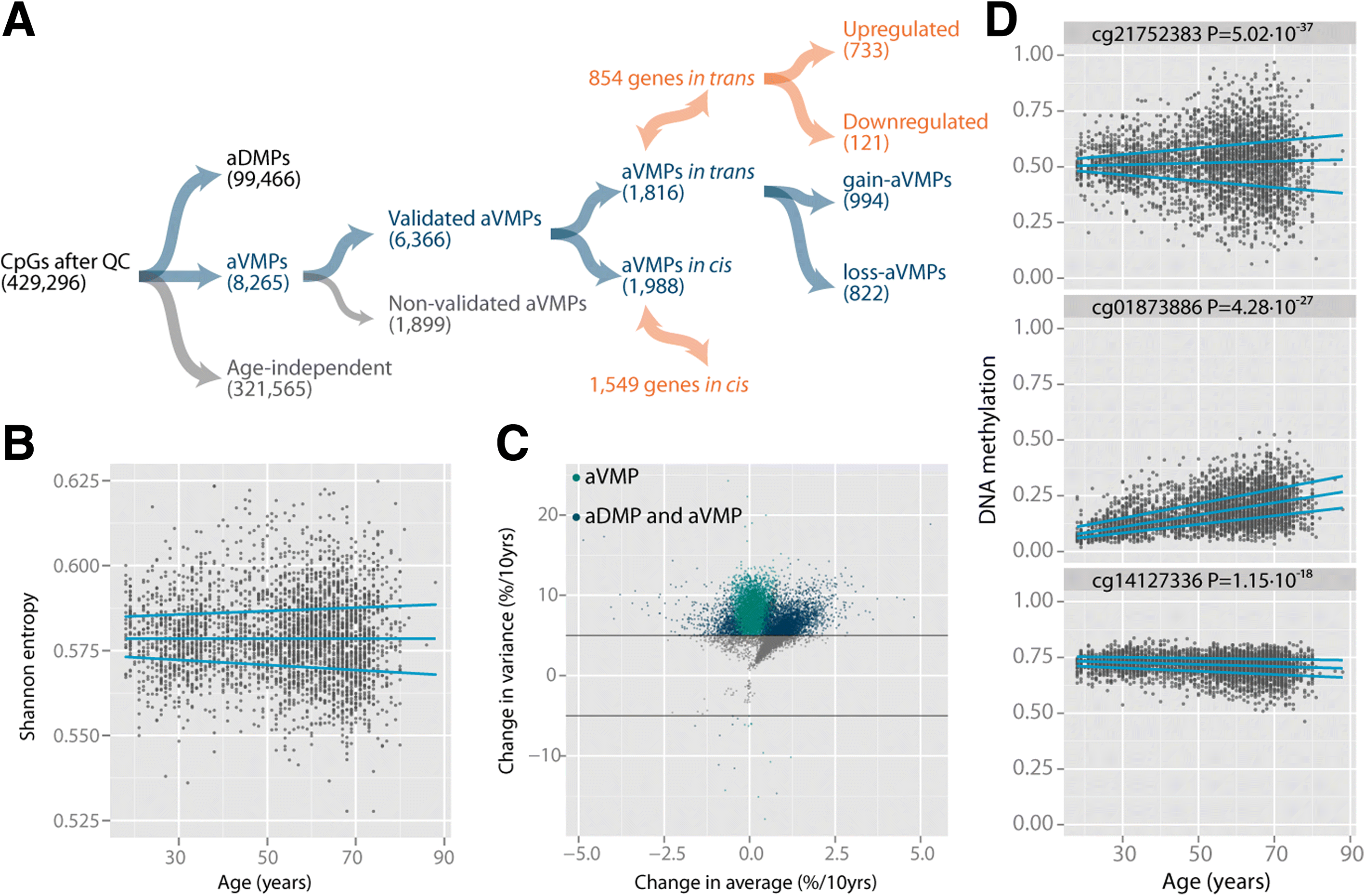
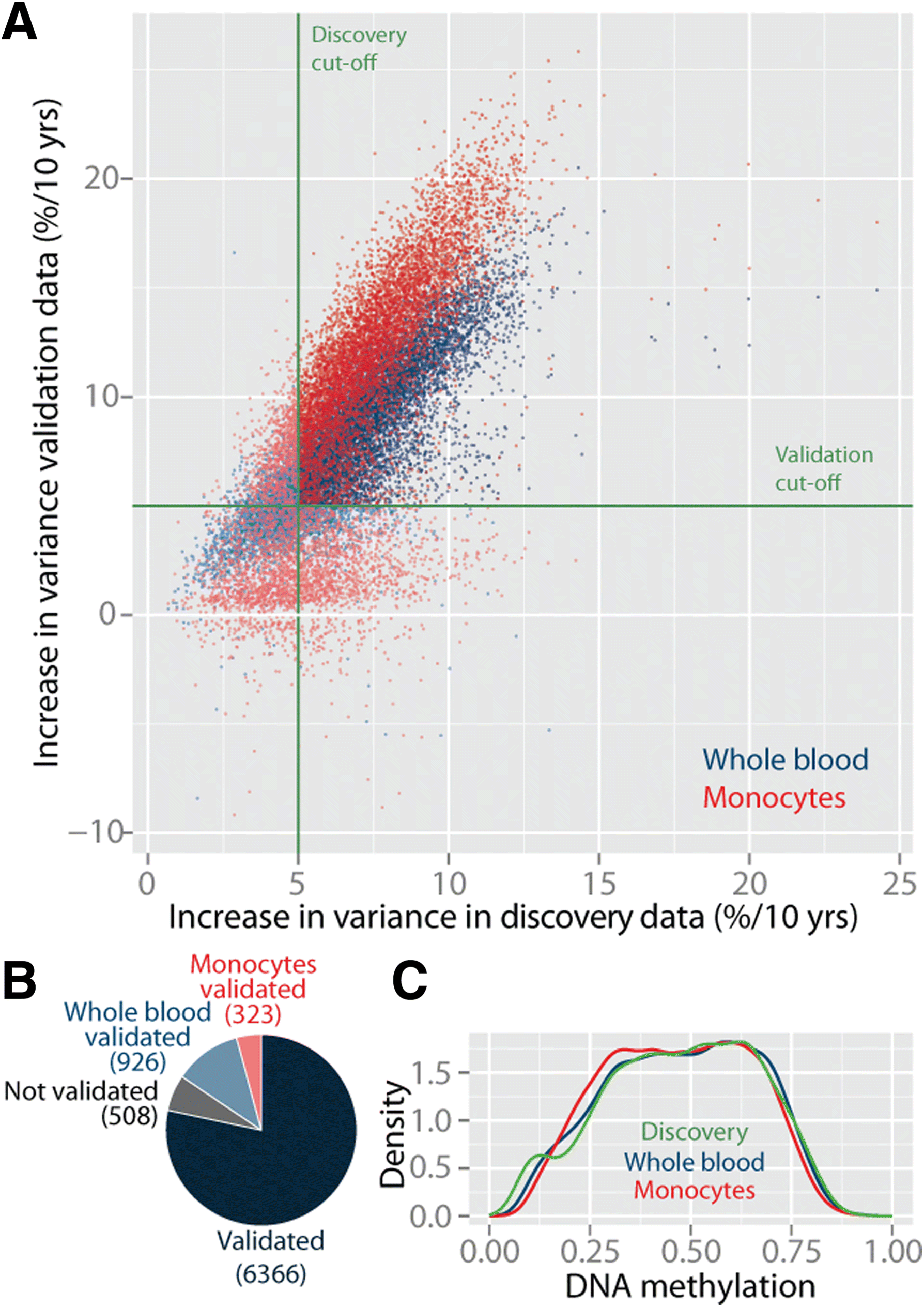
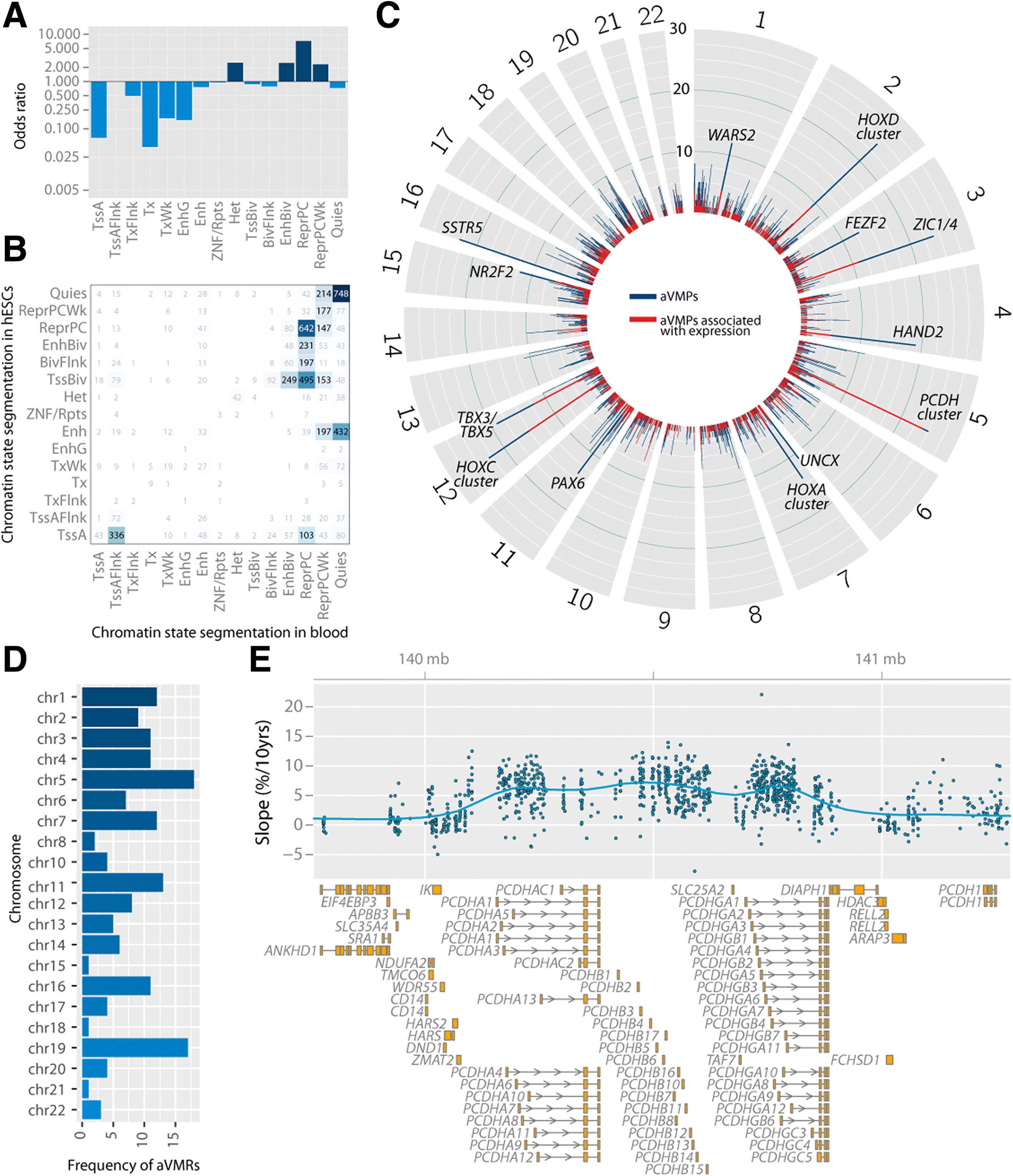
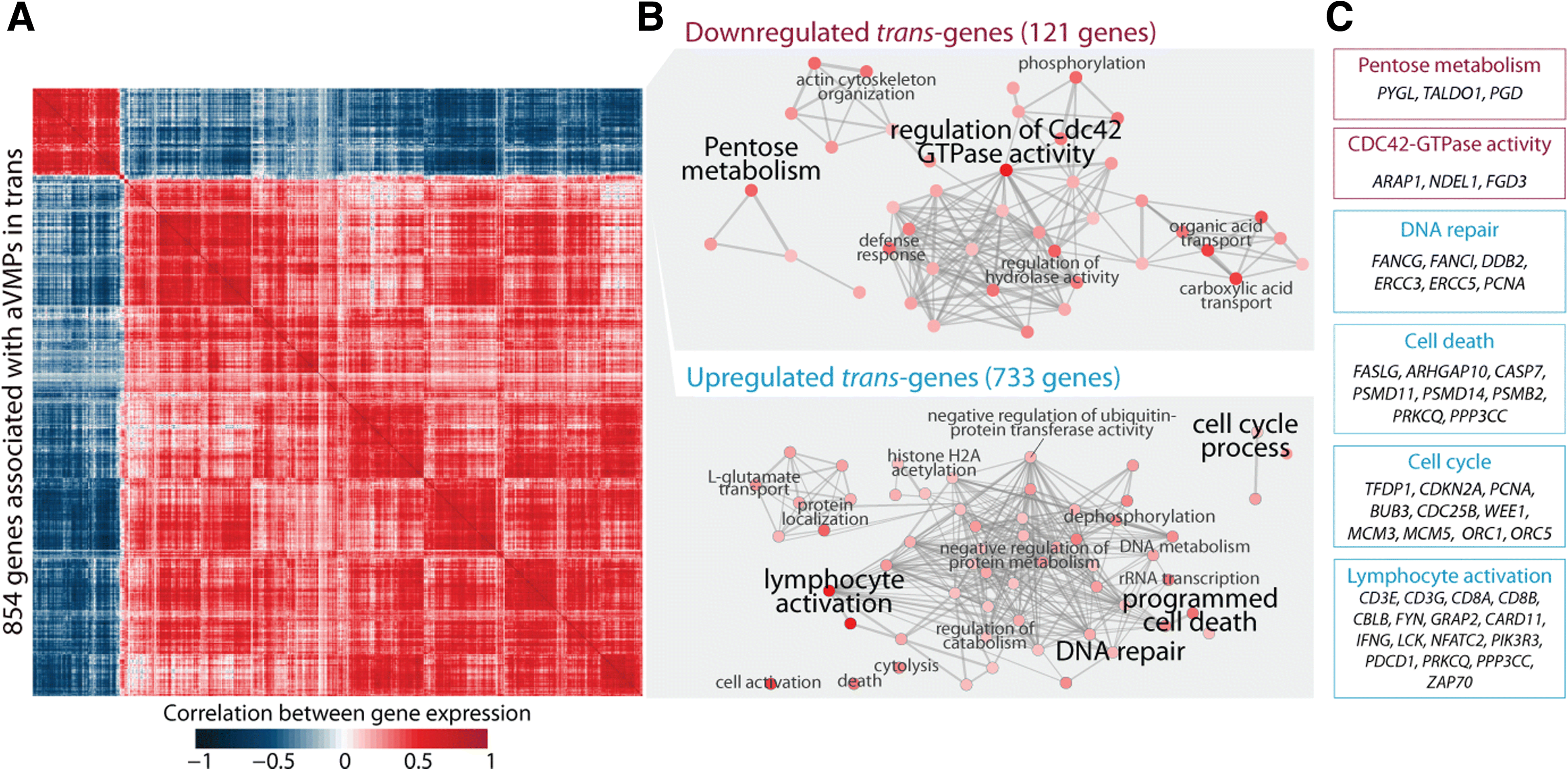
Results: Here, we report on the large-scale identification of 6366 age-related variably methylated positions (aVMPs) identified in 3295 whole blood DNA methylation profiles, 2044 of which have a matching RNA-seq gene expression profile. aVMPs are enriched at polycomb repressed regions and, accordingly, methylation at those positions is associated with the expression of genes encoding components of polycomb repressive complex 2 (PRC2) in trans. Further analysis revealed trans-associations for 1816 aVMPs with an additional 854 genes. These trans-associated aVMPs are characterized by either an age-related gain of methylation at CpG islands marked by PRC2 or a loss of methylation at enhancers. This distinct pattern extends to other tissues and multiple cancer types. Finally, genes associated with aVMPs in trans whose expression is variably upregulated with age (733 genes) play a key role in DNA repair and apoptosis, whereas downregulated aVMP-associated genes (121 genes) are mapped to defined pathways in cellular metabolism.
Conclusions: Our results link age-related changes in DNA methylation to fundamental mechanisms that are thought to drive human ageing.
Keywords: 450k; Ageing; DNA damage; DNA methylation; Variability.
Figures
Similar articles
-
Hypomethylation coordinates antagonistically with hypermethylation in cancer development: a case study of leukemia.Hum Genomics. 2016 Jul 25;10 Suppl 2(Suppl 2):18. doi: 10.1186/s40246-016-0071-5. Hum Genomics. 2016. PMID: 27461342 Free PMC article.
-
SIRT1 affects DNA methylation of polycomb group protein target genes, a hotspot of the epigenetic shift observed in ageing.Hum Genomics. 2015 Jun 24;9(1):14. doi: 10.1186/s40246-015-0036-0. Hum Genomics. 2015. PMID: 26104761 Free PMC article.
-
5'-Hydroxymethylcytosine Precedes Loss of CpG Methylation in Enhancers and Genes Undergoing Activation in Cardiomyocyte Maturation.PLoS One. 2016 Nov 16;11(11):e0166575. doi: 10.1371/journal.pone.0166575. eCollection 2016. PLoS One. 2016. PMID: 27851806 Free PMC article.
-
Mapping of Variable DNA Methylation Across Multiple Cell Types Defines a Dynamic Regulatory Landscape of the Human Genome.G3 (Bethesda). 2016 Apr 7;6(4):973-86. doi: 10.1534/g3.115.025437. G3 (Bethesda). 2016. PMID: 26888867 Free PMC article.
-
Genome-wide DNA methylome variation in two genetically distinct chicken lines using MethylC-seq.BMC Genomics. 2015 Oct 23;16:851. doi: 10.1186/s12864-015-2098-8. BMC Genomics. 2015. PMID: 26497311 Free PMC article.
Cited by
-
The immune factors driving DNA methylation variation in human blood.Nat Commun. 2022 Oct 6;13(1):5895. doi: 10.1038/s41467-022-33511-6. Nat Commun. 2022. PMID: 36202838 Free PMC article.
-
Characterizing epigenetic aging in an adult sickle cell disease cohort.Blood Adv. 2024 Jan 9;8(1):47-55. doi: 10.1182/bloodadvances.2023011188. Blood Adv. 2024. PMID: 37967379 Free PMC article.
-
Correlating blood-based DNA methylation markers and prostate cancer risk in African-American men.PLoS One. 2018 Sep 11;13(9):e0203322. doi: 10.1371/journal.pone.0203322. eCollection 2018. PLoS One. 2018. PMID: 30204798 Free PMC article.
-
DNA methylation associated with healthy aging of elderly twins.Geroscience. 2018 Dec;40(5-6):469-484. doi: 10.1007/s11357-018-0040-0. Epub 2018 Aug 22. Geroscience. 2018. PMID: 30136078 Free PMC article.
-
Enhanced epigenetic profiling of classical human monocytes reveals a specific signature of healthy aging in the DNA methylome.Nat Aging. 2021 Jan;1(1):124-141. doi: 10.1038/s43587-020-00002-6. Epub 2020 Nov 23. Nat Aging. 2021. PMID: 34796338 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

