The Comparative Toxicogenomics Database: update 2017
- PMID: 27651457
- PMCID: PMC5210612
- DOI: 10.1093/nar/gkw838
The Comparative Toxicogenomics Database: update 2017
Abstract
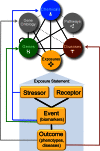
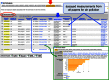
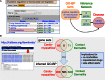
The Comparative Toxicogenomics Database (CTD; http://ctdbase.org/) provides information about interactions between chemicals and gene products, and their relationships to diseases. Core CTD content (chemical-gene, chemical-disease and gene-disease interactions manually curated from the literature) are integrated with each other as well as with select external datasets to generate expanded networks and predict novel associations. Today, core CTD includes more than 30.5 million toxicogenomic connections relating chemicals/drugs, genes/proteins, diseases, taxa, Gene Ontology (GO) annotations, pathways, and gene interaction modules. In this update, we report a 33% increase in our core data content since 2015, describe our new exposure module (that harmonizes exposure science information with core toxicogenomic data) and introduce a novel dataset of GO-disease inferences (that identify common molecular underpinnings for seemingly unrelated pathologies). These advancements centralize and contextualize real-world chemical exposures with molecular pathways to help scientists generate testable hypotheses in an effort to understand the etiology and mechanisms underlying environmentally influenced diseases.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Generating Gene Ontology-Disease Inferences to Explore Mechanisms of Human Disease at the Comparative Toxicogenomics Database.PLoS One. 2016 May 12;11(5):e0155530. doi: 10.1371/journal.pone.0155530. eCollection 2016. PLoS One. 2016. PMID: 27171405 Free PMC article.
-
The Comparative Toxicogenomics Database: update 2019.Nucleic Acids Res. 2019 Jan 8;47(D1):D948-D954. doi: 10.1093/nar/gky868. Nucleic Acids Res. 2019. PMID: 30247620 Free PMC article.
-
Comparative Toxicogenomics Database (CTD): update 2021.Nucleic Acids Res. 2021 Jan 8;49(D1):D1138-D1143. doi: 10.1093/nar/gkaa891. Nucleic Acids Res. 2021. PMID: 33068428 Free PMC article.
-
DiseaseMeth version 2.0: a major expansion and update of the human disease methylation database.Nucleic Acids Res. 2017 Jan 4;45(D1):D888-D895. doi: 10.1093/nar/gkw1123. Epub 2016 Nov 29. Nucleic Acids Res. 2017. PMID: 27899673 Free PMC article. Review.
-
The Comparative Toxicogenomics Database (CTD): a resource for comparative toxicological studies.J Exp Zool A Comp Exp Biol. 2006 Sep 1;305(9):689-92. doi: 10.1002/jez.a.307. J Exp Zool A Comp Exp Biol. 2006. PMID: 16902965 Free PMC article. Review.
Cited by
-
Identification of differentially expressed genes between mucinous adenocarcinoma and other adenocarcinoma of colorectal cancer using bioinformatics analysis.J Int Med Res. 2020 Aug;48(8):300060520949036. doi: 10.1177/0300060520949036. J Int Med Res. 2020. PMID: 32840168 Free PMC article.
-
The distributions of protein coding genes within chromatin domains in relation to human disease.Epigenetics Chromatin. 2019 Dec 5;12(1):72. doi: 10.1186/s13072-019-0317-2. Epigenetics Chromatin. 2019. PMID: 31805995 Free PMC article.
-
DigChem: Identification of disease-gene-chemical relationships from Medline abstracts.PLoS Comput Biol. 2019 May 15;15(5):e1007022. doi: 10.1371/journal.pcbi.1007022. eCollection 2019 May. PLoS Comput Biol. 2019. PMID: 31091224 Free PMC article.
-
Identification of Sex-Specific Transcriptome Responses to Polychlorinated Biphenyls (PCBs).Sci Rep. 2019 Jan 24;9(1):746. doi: 10.1038/s41598-018-37449-y. Sci Rep. 2019. PMID: 30679748 Free PMC article.
-
Natural products for infectious microbes and diseases: an overview of sources, compounds, and chemical diversities.Sci China Life Sci. 2022 Jun;65(6):1123-1145. doi: 10.1007/s11427-020-1959-5. Epub 2021 Oct 21. Sci China Life Sci. 2022. PMID: 34705221 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

