Antimicrobial peptide exposure selects for Staphylococcus aureus resistance to human defence peptides
- PMID: 27650186
- PMCID: PMC5161045
- DOI: 10.1093/jac/dkw381
Antimicrobial peptide exposure selects for Staphylococcus aureus resistance to human defence peptides
Abstract
Background: The clinical development of antimicrobial peptides (AMPs) is currently under evaluation to combat the rapid increase in MDR bacterial pathogens. However, many AMPs closely resemble components of the human innate immune system and the ramifications of prolonged bacterial exposure to AMPs are not fully understood.
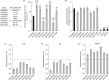
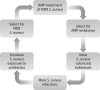
Objectives: We show that in vitro serial passage of a clinical USA300 MRSA strain in a host-mimicking environment containing host-derived AMPs results in the selection of stable AMP resistance.
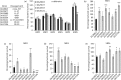
Methods: Serial passage experiments were conducted using steadily increasing concentrations of LL-37, PR-39 or wheat germ histones. WGS and proteomic analysis by MS were used to identify the molecular mechanism associated with increased tolerance of AMPs. AMP-resistant mutants were characterized by measuring in vitro fitness, AMP and antibiotic susceptibility, and virulence in a mouse model of sepsis.
Results: AMP-resistant Staphylococcus aureus mutants often displayed little to no fitness cost and caused invasive disease in mice. Further, this phenotype coincided with diminished susceptibility to both clinically prescribed antibiotics and human defence peptides.
Conclusions: These findings suggest that therapeutic use of AMPs could select for virulent mutants with cross-resistance to human innate immunity as well as antibiotic therapy. Thus, therapeutic use of AMPs and the implications of cross-resistance need to be carefully monitored and evaluated.
© The Author 2016. Published by Oxford University Press on behalf of the British Society for Antimicrobial Chemotherapy.
Figures


Similar articles
-
Mechanisms and consequences of bacterial resistance to antimicrobial peptides.Drug Resist Updat. 2016 May;26:43-57. doi: 10.1016/j.drup.2016.04.002. Epub 2016 Apr 20. Drug Resist Updat. 2016. PMID: 27180309 Review.
-
Community-acquired meticillin-resistant Staphylococcus aureus strain USA300 resists staphylococcal protein A modulation by antibiotics and antimicrobial peptides.Int J Antimicrob Agents. 2015 Jan;45(1):19-24. doi: 10.1016/j.ijantimicag.2014.08.009. Epub 2014 Oct 2. Int J Antimicrob Agents. 2015. PMID: 25450803
-
Antimicrobial activity of novel symmetrical antimicrobial peptides centered on a hydrophilic motif against resistant clinical isolates: in vitro and in vivo analyses.Microbiol Spectr. 2024 Nov 5;12(11):e0026524. doi: 10.1128/spectrum.00265-24. Epub 2024 Oct 9. Microbiol Spectr. 2024. PMID: 39382284 Free PMC article.
-
The antimicrobial peptide-sensing system aps of Staphylococcus aureus.Mol Microbiol. 2007 Dec;66(5):1136-47. doi: 10.1111/j.1365-2958.2007.05986.x. Epub 2007 Oct 24. Mol Microbiol. 2007. PMID: 17961141
-
Beyond conventional antibiotics for the future treatment of methicillin-resistant Staphylococcus aureus infections: two novel alternatives.FEMS Immunol Med Microbiol. 2012 Aug;65(3):399-412. doi: 10.1111/j.1574-695X.2012.00954.x. Epub 2012 Apr 4. FEMS Immunol Med Microbiol. 2012. PMID: 22409572 Review.
Cited by
-
Bacterial resistance to antibacterial agents: Mechanisms, control strategies, and implications for global health.Sci Total Environ. 2023 Feb 20;860:160461. doi: 10.1016/j.scitotenv.2022.160461. Epub 2022 Nov 23. Sci Total Environ. 2023. PMID: 36435256 Free PMC article. Review.
-
Incubation with a Complex Orange Essential Oil Leads to Evolved Mutants with Increased Resistance and Tolerance.Pharmaceuticals (Basel). 2020 Sep 9;13(9):239. doi: 10.3390/ph13090239. Pharmaceuticals (Basel). 2020. PMID: 32916977 Free PMC article.
-
Interrogation of Essentiality in the Reconstructed Haemophilus influenzae Metabolic Network Identifies Lipid Metabolism Antimicrobial Targets: Preclinical Evaluation of a FabH β-Ketoacyl-ACP Synthase Inhibitor.mSystems. 2022 Apr 26;7(2):e0145921. doi: 10.1128/msystems.01459-21. Epub 2022 Mar 16. mSystems. 2022. PMID: 35293791 Free PMC article.
-
In Vitro activity of novel glycopolymer against clinical isolates of multidrug-resistant Staphylococcus aureus.PLoS One. 2018 Jan 17;13(1):e0191522. doi: 10.1371/journal.pone.0191522. eCollection 2018. PLoS One. 2018. PMID: 29342216 Free PMC article.
-
Temporin B Forms Hetero-Oligomers with Temporin L, Modifies Its Membrane Activity, and Increases the Cooperativity of Its Antibacterial Pharmacodynamic Profile.Biochemistry. 2022 Jun 7;61(11):1029-1040. doi: 10.1021/acs.biochem.1c00762. Epub 2022 May 24. Biochemistry. 2022. PMID: 35609188 Free PMC article.
References
-
- WHO. Antimicrobial Resistance: Global Report on Surveillance. http://www.who.int/drugresistance/documents/surveillancereport/en/.
-
- van der Does AM, Bergman P, Agerberth B et al. . Induction of the human cathelicidin LL-37 as a novel treatment against bacterial infections. J Leukoc Biol 2012; 92: 735–42. - PubMed
-
- Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol 2006; 24: 1551–7. - PubMed
-
- Epand RM, Walker C, Epand RF et al. . Molecular mechanisms of membrane targeting antibiotics. Biochim Biophys Acta 2016; 1858: 980–7. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

