Amino Acids Regulate mTORC1 by an Obligate Two-step Mechanism
- PMID: 27587390
- PMCID: PMC5077182
- DOI: 10.1074/jbc.M116.732511
Amino Acids Regulate mTORC1 by an Obligate Two-step Mechanism
Erratum in
-
Correction: Amino acids regulate mTORC1 by an obligate two-step mechanism.J Biol Chem. 2020 Jun 26;295(26):8868. doi: 10.1074/jbc.AAC120.014360. J Biol Chem. 2020. PMID: 32591444 Free PMC article. No abstract available.
Abstract
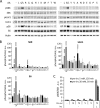
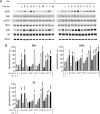
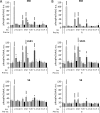
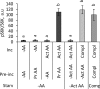
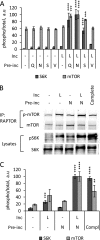
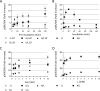
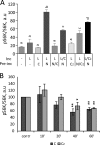
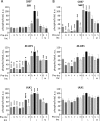
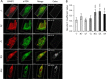
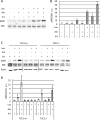
The mechanistic target of rapamycin complex 1 (mTORC1) coordinates cell growth with its nutritional, hormonal, energy, and stress status. Amino acids are critical regulators of mTORC1 that permit other inputs to mTORC1 activity. However, the roles of individual amino acids and their interactions in mTORC1 activation are not well understood. Here we demonstrate that activation of mTORC1 by amino acids includes two discrete and separable steps: priming and activation. Sensitizing mTORC1 activation by priming amino acids is a prerequisite for subsequent stimulation of mTORC1 by activating amino acids. Priming is achieved by a group of amino acids that includes l-asparagine, l-glutamine, l-threonine, l-arginine, l-glycine, l-proline, l-serine, l-alanine, and l-glutamic acid. The group of activating amino acids is dominated by l-leucine but also includes l-methionine, l-isoleucine, and l-valine. l-Cysteine predominantly inhibits priming but not the activating step. Priming and activating steps differ in their requirements for amino acid concentration and duration of treatment. Priming and activating amino acids use mechanisms that are distinct both from each other and from growth factor signaling. Neither step requires intact tuberous sclerosis complex of proteins to activate mTORC1. Concerted action of priming and activating amino acids is required to localize mTORC1 to lysosomes and achieve its activation.
Keywords: S6 kinase; amino acid; cell signaling; lysosome; mTOR complex (mTORC); mammalian target of rapamycin (mTOR); protein kinase.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
Similar articles
-
Structure-activity relations of leucine derivatives reveal critical moieties for cellular uptake and activation of mTORC1-mediated signaling.Amino Acids. 2016 Apr;48(4):1045-1058. doi: 10.1007/s00726-015-2158-z. Epub 2016 Jan 2. Amino Acids. 2016. PMID: 26724922
-
Amino acids and mTORC1: from lysosomes to disease.Trends Mol Med. 2012 Sep;18(9):524-33. doi: 10.1016/j.molmed.2012.05.007. Epub 2012 Jun 28. Trends Mol Med. 2012. PMID: 22749019 Free PMC article. Review.
-
Requirement for lysosomal localization of mTOR for its activation differs between leucine and other amino acids.Cell Signal. 2014 Sep;26(9):1918-27. doi: 10.1016/j.cellsig.2014.04.019. Epub 2014 May 2. Cell Signal. 2014. PMID: 24793303
-
Nutrient signaling to mTOR and cell growth.Trends Biochem Sci. 2013 May;38(5):233-42. doi: 10.1016/j.tibs.2013.01.004. Epub 2013 Mar 1. Trends Biochem Sci. 2013. PMID: 23465396 Free PMC article. Review.
-
mTORC1 Activator SLC38A9 Is Required to Efflux Essential Amino Acids from Lysosomes and Use Protein as a Nutrient.Cell. 2017 Oct 19;171(3):642-654.e12. doi: 10.1016/j.cell.2017.09.046. Cell. 2017. PMID: 29053970 Free PMC article.
Cited by
-
Metabolic compensation activates pro-survival mTORC1 signaling upon 3-phosphoglycerate dehydrogenase inhibition in osteosarcoma.Cell Rep. 2021 Jan 26;34(4):108678. doi: 10.1016/j.celrep.2020.108678. Cell Rep. 2021. PMID: 33503424 Free PMC article.
-
Targeting BCAT1 Combined with α-Ketoglutarate Triggers Metabolic Synthetic Lethality in Glioblastoma.Cancer Res. 2022 Jul 5;82(13):2388-2402. doi: 10.1158/0008-5472.CAN-21-3868. Cancer Res. 2022. PMID: 35499760 Free PMC article.
-
LKB1 specifies neural crest cell fates through pyruvate-alanine cycling.Sci Adv. 2019 Jul 17;5(7):eaau5106. doi: 10.1126/sciadv.aau5106. eCollection 2019 Jul. Sci Adv. 2019. PMID: 31328154 Free PMC article.
-
Glutamine synthetase limits β-catenin-mutated liver cancer growth by maintaining nitrogen homeostasis and suppressing mTORC1.J Clin Invest. 2022 Dec 15;132(24):e161408. doi: 10.1172/JCI161408. J Clin Invest. 2022. PMID: 36256480 Free PMC article.
-
Interplays of AMPK and TOR in Autophagy Regulation in Yeast.Cells. 2023 Feb 4;12(4):519. doi: 10.3390/cells12040519. Cells. 2023. PMID: 36831186 Free PMC article. Review.
References
-
- Ma X. M., Yoon S.-O., Richardson C. J., Jülich K., and Blenis J. (2008) SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell 133, 303–313 - PubMed
-
- Gingras A. C., Raught B., and Sonenberg N. (2004) mTOR signaling to translation. Curr. Top. Microbiol. Immunol. 279, 169–197 - PubMed
-
- Guertin D. A., and Sabatini D. M. (2007) Defining the role of mTOR in cancer. Cancer Cell 12, 9–22 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

