Notch activation drives adipocyte dedifferentiation and tumorigenic transformation in mice
- PMID: 27573812
- PMCID: PMC5030803
- DOI: 10.1084/jem.20160157
Notch activation drives adipocyte dedifferentiation and tumorigenic transformation in mice
Abstract
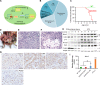
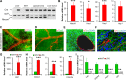
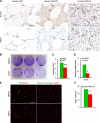
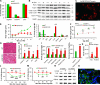
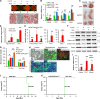
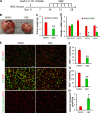
Liposarcomas (LPSs) are the most common soft-tissue cancer. Because of the lack of animal models, the cellular origin and molecular regulation of LPS remain unclear. Here, we report that mice with adipocyte-specific activation of Notch signaling (Ad/N1ICD) develop LPS with complete penetrance. Lineage tracing confirms the adipocyte origin of Ad/N1ICD LPS. The Ad/N1ICD LPS resembles human dedifferentiated LPS in histological appearance, anatomical localization, and gene expression signature. Before transformation, Ad/N1ICD adipocytes undergo dedifferentiation that leads to lipodystrophy and metabolic dysfunction. Although concomitant Pten deletion normalizes the glucose metabolism of Ad/N1ICD mice, it dramatically accelerates the LPS prognosis and malignancy. Transcriptomes and lipidomics analyses indicate that Notch activation suppresses lipid metabolism pathways that supply ligands to Pparγ, the master regulator of adipocyte homeostasis. Accordingly, synthetic Pparγ ligand supplementation induces redifferentiation of Ad/N1ICD adipocytes and tumor cells, and prevents LPS development in Ad/N1ICD mice. Importantly, the Notch target HES1 is abundantly expressed in human LPS, and Notch inhibition suppresses the growth of human dedifferentiated LPS xenografts. Collectively, ectopic Notch activation is sufficient to induce dedifferentiation and tumorigenic transformation of mature adipocytes in mouse.
© 2016 Bi et al.
Figures



Similar articles
-
Sustained activation of notch signaling maintains tumor-initiating cells in a murine model of liposarcoma.Cancer Lett. 2020 Dec 1;494:27-39. doi: 10.1016/j.canlet.2020.08.029. Epub 2020 Aug 28. Cancer Lett. 2020. PMID: 32866607 Free PMC article.
-
Novel dedifferentiated liposarcoma xenograft models reveal PTEN down-regulation as a malignant signature and response to PI3K pathway inhibition.Am J Pathol. 2013 Apr;182(4):1400-11. doi: 10.1016/j.ajpath.2013.01.002. Epub 2013 Feb 12. Am J Pathol. 2013. PMID: 23416162 Free PMC article.
-
Phosphatase and tensin homolog deleted on chromosome 10 suppression is an important process in peroxisome proliferator-activated receptor-gamma signaling in adipocytes and myotubes.Mol Pharmacol. 2007 Jun;71(6):1554-62. doi: 10.1124/mol.106.031948. Epub 2007 Mar 2. Mol Pharmacol. 2007. PMID: 17337625
-
The NOTCH pathway in β-cell growth and differentiation.Vitam Horm. 2014;95:391-405. doi: 10.1016/B978-0-12-800174-5.00015-6. Vitam Horm. 2014. PMID: 24559926 Review.
-
Roles of Notch Signaling in Adipocyte Progenitor Cells and Mature Adipocytes.J Cell Physiol. 2017 Jun;232(6):1258-1261. doi: 10.1002/jcp.25697. Epub 2017 Jan 5. J Cell Physiol. 2017. PMID: 27869309 Review.
Cited by
-
Ascl2 inhibits myogenesis by antagonizing the transcriptional activity of myogenic regulatory factors.Development. 2017 Jan 15;144(2):235-247. doi: 10.1242/dev.138099. Epub 2016 Dec 19. Development. 2017. PMID: 27993983 Free PMC article.
-
Roles of transducin-like enhancer of split (TLE) family proteins in tumorigenesis and immune regulation.Front Cell Dev Biol. 2022 Nov 11;10:1010639. doi: 10.3389/fcell.2022.1010639. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36438567 Free PMC article. Review.
-
Distinct roles for Notch1 and Notch3 in human adipose-derived stem/stromal cell adipogenesis.Mol Biol Rep. 2020 Nov;47(11):8439-8450. doi: 10.1007/s11033-020-05884-8. Epub 2020 Oct 6. Mol Biol Rep. 2020. PMID: 33021719
-
Emerging role of tumor cell plasticity in modifying therapeutic response.Signal Transduct Target Ther. 2020 Oct 7;5(1):228. doi: 10.1038/s41392-020-00313-5. Signal Transduct Target Ther. 2020. PMID: 33028808 Free PMC article. Review.
-
Fusobacterium nucleatum Promotes Colorectal Cancer Cell to Acquire Stem Cell-Like Features by Manipulating Lipid Droplet-Mediated Numb Degradation.Adv Sci (Weinh). 2022 Apr;9(12):e2105222. doi: 10.1002/advs.202105222. Epub 2022 Feb 15. Adv Sci (Weinh). 2022. PMID: 35170250 Free PMC article.
References
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

