A Role for Nuclear F-Actin Induction in Human Cytomegalovirus Nuclear Egress
- PMID: 27555312
- PMCID: PMC4999551
- DOI: 10.1128/mBio.01254-16
A Role for Nuclear F-Actin Induction in Human Cytomegalovirus Nuclear Egress
Abstract
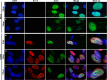
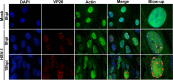
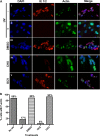
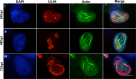
Herpesviruses, which include important pathogens, remodel the host cell nucleus to facilitate infection. This remodeling includes the formation of structures called replication compartments (RCs) in which herpesviruses replicate their DNA. During infection with the betaherpesvirus, human cytomegalovirus (HCMV), viral DNA synthesis occurs at the periphery of RCs within the nuclear interior, after which assembled capsids must reach the inner nuclear membrane (INM) for translocation to the cytoplasm (nuclear egress). The processes that facilitate movement of HCMV capsids to the INM during nuclear egress are unknown. Although an actin-based mechanism of alphaherpesvirus capsid trafficking to the INM has been proposed, it is controversial. Here, using a fluorescently-tagged, nucleus-localized actin-binding peptide, we show that HCMV, but not herpes simplex virus 1, strongly induced nuclear actin filaments (F-actin) in human fibroblasts. Based on studies using UV inactivation and inhibitors, this induction depended on viral gene expression. Interestingly, by 24 h postinfection, nuclear F-actin formed thicker structures that appeared by super-resolution microscopy to be bundles of filaments. Later in infection, nuclear F-actin primarily localized along the RC periphery and between the RC periphery and the nuclear rim. Importantly, a drug that depolymerized nuclear F-actin caused defects in production of infectious virus, capsid accumulation in the cytoplasm, and capsid localization near the nuclear rim, without decreasing capsid accumulation in the nucleus. Thus, our results suggest that for at least one herpesvirus, nuclear F-actin promotes capsid movement to the nuclear periphery and nuclear egress. We discuss our results in terms of competing models for these processes.
Importance: The mechanisms underlying herpesvirus nuclear egress have not been fully determined. In particular, how newly assembled capsids move to the inner nuclear membrane for envelopment is uncertain and controversial. In this study, we show that HCMV, an important human pathogen, induces actin filaments in the nuclei of infected cells and that an inhibitor of nuclear F-actin impairs nuclear egress and capsid localization toward the nuclear periphery. Herpesviruses are widespread pathogens that cause or contribute to an array of human diseases. A better understanding of how herpesvirus capsids traffic in the nucleus may uncover novel targets for antiviral intervention and elucidate aspects of the nuclear cytoskeleton, about which little is known.
Copyright © 2016 Wilkie et al.
Figures

Similar articles
-
A Role for Myosin Va in Human Cytomegalovirus Nuclear Egress.J Virol. 2018 Feb 26;92(6):e01849-17. doi: 10.1128/JVI.01849-17. Print 2018 Mar 15. J Virol. 2018. PMID: 29298889 Free PMC article.
-
Nuclear herpesvirus capsid motility is not dependent on F-actin.mBio. 2014 Oct 7;5(5):e01909-14. doi: 10.1128/mBio.01909-14. mBio. 2014. PMID: 25293761 Free PMC article.
-
Human Cytomegalovirus Nuclear Capsids Associate with the Core Nuclear Egress Complex and the Viral Protein Kinase pUL97.Viruses. 2018 Jan 13;10(1):35. doi: 10.3390/v10010035. Viruses. 2018. PMID: 29342872 Free PMC article.
-
The human cytomegalovirus nuclear egress complex unites multiple functions: Recruitment of effectors, nuclear envelope rearrangement, and docking to nuclear capsids.Rev Med Virol. 2017 Jul;27(4). doi: 10.1002/rmv.1934. Epub 2017 Jun 30. Rev Med Virol. 2017. PMID: 28664574 Review.
-
Getting to and through the inner nuclear membrane during herpesvirus nuclear egress.Curr Opin Cell Biol. 2017 Jun;46:9-16. doi: 10.1016/j.ceb.2016.12.007. Epub 2017 Jan 10. Curr Opin Cell Biol. 2017. PMID: 28086162 Free PMC article. Review.
Cited by
-
Human Cytomegalovirus Nuclear Egress Complex Subunit, UL53, Associates with Capsids and Myosin Va, but Is Not Important for Capsid Localization towards the Nuclear Periphery.Viruses. 2022 Feb 26;14(3):479. doi: 10.3390/v14030479. Viruses. 2022. PMID: 35336886 Free PMC article.
-
A Role for Myosin Va in Human Cytomegalovirus Nuclear Egress.J Virol. 2018 Feb 26;92(6):e01849-17. doi: 10.1128/JVI.01849-17. Print 2018 Mar 15. J Virol. 2018. PMID: 29298889 Free PMC article.
-
[Nucleus translocation of membrane/cytoplasm proteins in tumor cells].Zhejiang Da Xue Xue Bao Yi Xue Ban. 2019 May 25;48(3):318-325. doi: 10.3785/j.issn.1008-9292.2019.06.13. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2019. PMID: 31496165 Free PMC article. Chinese.
-
Nuclear Cytoskeleton in Virus Infection.Int J Mol Sci. 2022 Jan 5;23(1):578. doi: 10.3390/ijms23010578. Int J Mol Sci. 2022. PMID: 35009004 Free PMC article. Review.
-
Linking indirect effects of cytomegalovirus in transplantation to modulation of monocyte innate immune function.Sci Adv. 2020 Apr 22;6(17):eaax9856. doi: 10.1126/sciadv.aax9856. eCollection 2020 Apr. Sci Adv. 2020. PMID: 32494628 Free PMC article.
References
-
- Mocarski ES, Shenk T, Griffiths P, Pass R. 2013. Cytomegaloviruses, p 1960–2014. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
