Nuclear Perilipin 5 integrates lipid droplet lipolysis with PGC-1α/SIRT1-dependent transcriptional regulation of mitochondrial function
- PMID: 27554864
- PMCID: PMC4999519
- DOI: 10.1038/ncomms12723
Nuclear Perilipin 5 integrates lipid droplet lipolysis with PGC-1α/SIRT1-dependent transcriptional regulation of mitochondrial function
Abstract
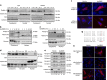
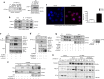
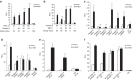
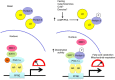
Dysfunctional cellular lipid metabolism contributes to common chronic human diseases, including type 2 diabetes, obesity, fatty liver disease and diabetic cardiomyopathy. How cells balance lipid storage and mitochondrial oxidative capacity is poorly understood. Here we identify the lipid droplet protein Perilipin 5 as a catecholamine-triggered interaction partner of PGC-1α. We report that during catecholamine-stimulated lipolysis, Perilipin 5 is phosphorylated by protein kinase A and forms transcriptional complexes with PGC-1α and SIRT1 in the nucleus. Perilipin 5 promotes PGC-1α co-activator function by disinhibiting SIRT1 deacetylase activity. We show by gain-and-loss of function studies in cells that nuclear Perilipin 5 promotes transcription of genes that mediate mitochondrial biogenesis and oxidative function. We propose that Perilipin 5 is an important molecular link that couples the coordinated catecholamine activation of the PKA pathway and of lipid droplet lipolysis with transcriptional regulation to promote efficient fatty acid catabolism and prevent mitochondrial dysfunction.
Figures



References
-
- Ducharme N. A. & Bickel P. E. Lipid droplets in lipogenesis and lipolysis. Endocrinology 149, 942–949 (2008). - PubMed
-
- Koves T. R. et al.. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 7, 45–56 (2008). - PubMed
-
- Koves T. R. et al.. Peroxisome proliferator-activated receptor-gamma co-activator 1alpha-mediated metabolic remodeling of skeletal myocytes mimics exercise training and reverses lipid-induced mitochondrial inefficiency. J. Biol. Chem. 280, 33588–33598 (2005). - PubMed
-
- Wu Z. et al.. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98, 115–124 (1999). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

