Ferroptosis is an autophagic cell death process
- PMID: 27514700
- PMCID: PMC5034113
- DOI: 10.1038/cr.2016.95
Ferroptosis is an autophagic cell death process
Abstract
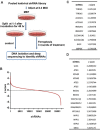
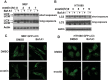
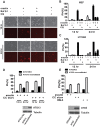
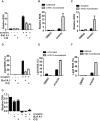
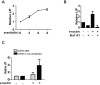
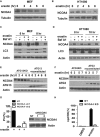
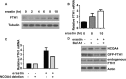
Ferroptosis is an iron-dependent form of regulated necrosis. It is implicated in various human diseases, including ischemic organ damage and cancer. Here, we report the crucial role of autophagy, particularly autophagic degradation of cellular iron storage proteins (a process known as ferritinophagy), in ferroptosis. Using RNAi screening coupled with subsequent genetic analysis, we identified multiple autophagy-related genes as positive regulators of ferroptosis. Ferroptosis induction led to autophagy activation and consequent degradation of ferritin and ferritinophagy cargo receptor NCOA4. Consistently, inhibition of ferritinophagy by blockage of autophagy or knockdown of NCOA4 abrogated the accumulation of ferroptosis-associated cellular labile iron and reactive oxygen species, as well as eventual ferroptotic cell death. Therefore, ferroptosis is an autophagic cell death process, and NCOA4-mediated ferritinophagy supports ferroptosis by controlling cellular iron homeostasis.
Figures
Similar articles
-
Autophagy promotes ferroptosis by degradation of ferritin.Autophagy. 2016 Aug 2;12(8):1425-8. doi: 10.1080/15548627.2016.1187366. Epub 2016 May 31. Autophagy. 2016. PMID: 27245739 Free PMC article.
-
NCOA4-mediated ferritinophagy promotes ferroptosis induced by erastin, but not by RSL3 in HeLa cells.Biochim Biophys Acta Mol Cell Res. 2021 Feb;1868(2):118913. doi: 10.1016/j.bbamcr.2020.118913. Epub 2020 Nov 25. Biochim Biophys Acta Mol Cell Res. 2021. PMID: 33245979
-
The Role of NCOA4-Mediated Ferritinophagy in Ferroptosis.Adv Exp Med Biol. 2021;1301:41-57. doi: 10.1007/978-3-030-62026-4_4. Adv Exp Med Biol. 2021. PMID: 34370287 Review.
-
Ferroptosis: Role of lipid peroxidation, iron and ferritinophagy.Biochim Biophys Acta Gen Subj. 2017 Aug;1861(8):1893-1900. doi: 10.1016/j.bbagen.2017.05.019. Epub 2017 May 24. Biochim Biophys Acta Gen Subj. 2017. PMID: 28552631 Review.
-
Polybrominated Diphenyl Ethers Quinone Induces NCOA4-Mediated Ferritinophagy through Selectively Autophagic Degradation of Ferritin.Chem Res Toxicol. 2019 Dec 16;32(12):2509-2516. doi: 10.1021/acs.chemrestox.9b00350. Epub 2019 Nov 11. Chem Res Toxicol. 2019. PMID: 31687807
Cited by
-
Ferroptosis of Endothelial Cells in Vascular Diseases.Nutrients. 2022 Oct 26;14(21):4506. doi: 10.3390/nu14214506. Nutrients. 2022. PMID: 36364768 Free PMC article. Review.
-
Noncoding RNAs regulating ferroptosis in cardiovascular diseases: novel roles and therapeutic strategies.Mol Cell Biochem. 2024 Nov;479(11):2827-2841. doi: 10.1007/s11010-023-04895-w. Epub 2023 Dec 8. Mol Cell Biochem. 2024. PMID: 38064139 Free PMC article. Review.
-
Natural products targeting ferroptosis pathways in cancer therapy (Review).Oncol Rep. 2024 Sep;52(3):123. doi: 10.3892/or.2024.8782. Epub 2024 Jul 26. Oncol Rep. 2024. PMID: 39054952 Free PMC article. Review.
-
Revisiting the potential of regulated cell death in glioma treatment: a focus on autophagy-dependent cell death, anoikis, ferroptosis, cuproptosis, pyroptosis, immunogenic cell death, and the crosstalk between them.Front Oncol. 2024 Aug 9;14:1397863. doi: 10.3389/fonc.2024.1397863. eCollection 2024. Front Oncol. 2024. PMID: 39184045 Free PMC article. Review.
-
Inhibitory effect of hydnocarpin D on T-cell acute lymphoblastic leukemia via induction of autophagy-dependent ferroptosis.Exp Biol Med (Maywood). 2021 Jul;246(13):1541-1553. doi: 10.1177/15353702211004870. Epub 2021 Apr 29. Exp Biol Med (Maywood). 2021. PMID: 33926261 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

