Long and Short Isoforms of the Human Cytomegalovirus UL138 Protein Silence IE Transcription and Promote Latency
- PMID: 27512069
- PMCID: PMC5044833
- DOI: 10.1128/JVI.01547-16
Long and Short Isoforms of the Human Cytomegalovirus UL138 Protein Silence IE Transcription and Promote Latency
Abstract
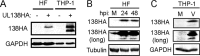
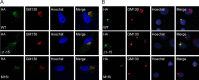
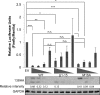
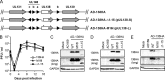
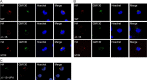
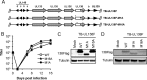
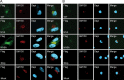
The UL133-138 locus present in clinical strains of human cytomegalovirus (HCMV) encodes proteins required for latency and reactivation in CD34(+) hematopoietic progenitor cells and virion maturation in endothelial cells. The encoded proteins form multiple homo- and hetero-interactions and localize within secretory membranes. One of these genes, UL136 gene, is expressed as at least five different protein isoforms with overlapping and unique functions. Here we show that another gene from this locus, the UL138 gene, also generates more than one protein isoform. A long form of UL138 (pUL138-L) initiates translation from codon 1, possesses an amino-terminal signal sequence, and is a type one integral membrane protein. Here we identify a short protein isoform (pUL138-S) initiating from codon 16 that displays a subcellular localization similar to that of pUL138-L. Reporter, short-term transcription, and long-term virus production assays revealed that both pUL138-L and pUL138-S are able to suppress major immediate early (IE) gene transcription and the generation of infectious virions in cells in which HCMV latency is studied. The long form appears to be more potent at silencing IE transcription shortly after infection, while the short form seems more potent at restricting progeny virion production at later times, indicating that both isoforms of UL138 likely cooperate to promote HCMV latency.
Importance: Latency allows herpesviruses to persist for the lives of their hosts in the face of effective immune control measures for productively infected cells. Controlling latent reservoirs is an attractive antiviral approach complicated by knowledge deficits for how latently infected cells are established, maintained, and reactivated. This is especially true for betaherpesviruses. The functional consequences of HCMV UL138 protein expression during latency include repression of viral IE1 transcription and suppression of virus replication. Here we show that short and long isoforms of UL138 exist and can themselves support latency but may do so in temporally distinct manners. Understanding the complexity of gene expression and its impact on latency is important for considering potential antivirals targeting latent reservoirs.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
The Role of the Human Cytomegalovirus UL133-UL138 Gene Locus in Latency and Reactivation.Viruses. 2020 Jul 1;12(7):714. doi: 10.3390/v12070714. Viruses. 2020. PMID: 32630219 Free PMC article. Review.
-
Antagonistic determinants controlling replicative and latent states of human cytomegalovirus infection.J Virol. 2014 Jun;88(11):5987-6002. doi: 10.1128/JVI.03506-13. Epub 2014 Mar 12. J Virol. 2014. PMID: 24623432 Free PMC article.
-
Complex Interplay of the UL136 Isoforms Balances Cytomegalovirus Replication and Latency.mBio. 2016 Mar 1;7(2):e01986. doi: 10.1128/mBio.01986-15. mBio. 2016. PMID: 26933055 Free PMC article.
-
Characterization of a novel Golgi apparatus-localized latency determinant encoded by human cytomegalovirus.J Virol. 2009 Jun;83(11):5615-29. doi: 10.1128/JVI.01989-08. Epub 2009 Mar 18. J Virol. 2009. PMID: 19297488 Free PMC article.
-
Bright and Early: Inhibiting Human Cytomegalovirus by Targeting Major Immediate-Early Gene Expression or Protein Function.Viruses. 2020 Jan 16;12(1):110. doi: 10.3390/v12010110. Viruses. 2020. PMID: 31963209 Free PMC article. Review.
Cited by
-
The Role of the Human Cytomegalovirus UL133-UL138 Gene Locus in Latency and Reactivation.Viruses. 2020 Jul 1;12(7):714. doi: 10.3390/v12070714. Viruses. 2020. PMID: 32630219 Free PMC article. Review.
-
NFκB and Cyclic AMP Response Element Sites Mediate the Valproic Acid and UL138 Responsiveness of the Human Cytomegalovirus Major Immediate Early Enhancer and Promoter.J Virol. 2023 Mar 30;97(3):e0002923. doi: 10.1128/jvi.00029-23. Epub 2023 Mar 1. J Virol. 2023. PMID: 36856444 Free PMC article.
-
The Membrane-Spanning Peptide and Acidic Cluster Dileucine Sorting Motif of UL138 Are Required To Downregulate MRP1 Drug Transporter Function in Human Cytomegalovirus-Infected Cells.J Virol. 2019 May 15;93(11):e00430-19. doi: 10.1128/JVI.00430-19. Print 2019 Jun 1. J Virol. 2019. PMID: 30894470 Free PMC article.
-
The Cytomegalovirus Protein Kinase pUL97:Host Interactions, Regulatory Mechanisms and Antiviral Drug Targeting.Microorganisms. 2020 Apr 4;8(4):515. doi: 10.3390/microorganisms8040515. Microorganisms. 2020. PMID: 32260430 Free PMC article. Review.
-
The Golgi sorting motifs of human cytomegalovirus UL138 are not required for latency maintenance.Virus Res. 2019 Sep;270:197646. doi: 10.1016/j.virusres.2019.197646. Epub 2019 Jun 28. Virus Res. 2019. PMID: 31260705 Free PMC article.
References
-
- Britt W. 2008. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr Top Microbiol Immunol 325:417–470. - PubMed
-
- Mocarski ES, Shenk T, Pass RF. 2007. Cytomegaloviruses, p 2701–2673. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott, Williams & Wilkins, Philadelphia, PA.
-
- Sinzger C, Digel M, Jahn G. 2008. Cytomegalovirus cell tropism. Curr Top Microbiol Immunol 325:63–83. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

