Evaluating Clonal Expansion of HIV-Infected Cells: Optimization of PCR Strategies to Predict Clonality
- PMID: 27494508
- PMCID: PMC4975415
- DOI: 10.1371/journal.ppat.1005689
Evaluating Clonal Expansion of HIV-Infected Cells: Optimization of PCR Strategies to Predict Clonality
Abstract
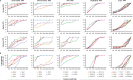
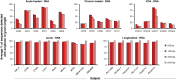
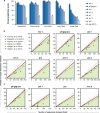
In HIV-infected individuals receiving suppressive antiretroviral therapy, the virus persists indefinitely in a reservoir of latently infected cells. The proliferation of these cells may contribute to the stability of the reservoir and thus to the lifelong persistence of HIV-1 in infected individuals. Because the HIV-1 replication process is highly error-prone, the detection of identical viral genomes in distinct host cells provides evidence for the clonal expansion of infected cells. We evaluated alignments of unique, near-full-length HIV-1 sequences to determine the relationship between clonality in a short region and clonality in the full genome. Although it is common to amplify and sequence short, subgenomic regions of the viral genome for phylogenetic analysis, we show that sequence identity of these amplicons does not guarantee clonality across the full viral genome. We show that although longer amplicons capture more diversity, no subgenomic region can recapitulate the diversity of full viral genomes. Consequently, some identical subgenomic amplicons should be expected even from the analysis of completely unique viral genomes, and the presence of identical amplicons alone is not proof of clonally expanded HIV-1. We present a method for evaluating evidence of clonal expansion in the context of these findings.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
The role of integration and clonal expansion in HIV infection: live long and prosper.Retrovirology. 2018 Oct 23;15(1):71. doi: 10.1186/s12977-018-0448-8. Retrovirology. 2018. PMID: 30352600 Free PMC article. Review.
-
HIV Diversity and Genetic Compartmentalization in Blood and Testes during Suppressive Antiretroviral Therapy.J Virol. 2019 Aug 13;93(17):e00755-19. doi: 10.1128/JVI.00755-19. Print 2019 Sep 1. J Virol. 2019. PMID: 31189714 Free PMC article.
-
Dynamics and mechanisms of clonal expansion of HIV-1-infected cells in a humanized mouse model.Sci Rep. 2017 Jul 31;7(1):6913. doi: 10.1038/s41598-017-07307-4. Sci Rep. 2017. PMID: 28761140 Free PMC article.
-
CD161+ CD4+ T Cells Harbor Clonally Expanded Replication-Competent HIV-1 in Antiretroviral Therapy-Suppressed Individuals.mBio. 2019 Oct 8;10(5):e02121-19. doi: 10.1128/mBio.02121-19. mBio. 2019. PMID: 31594817 Free PMC article.
-
Clonal Expansion of Human Immunodeficiency Virus-Infected Cells and Human Immunodeficiency Virus Persistence During Antiretroviral Therapy.J Infect Dis. 2017 Mar 15;215(suppl_3):S119-S127. doi: 10.1093/infdis/jiw636. J Infect Dis. 2017. PMID: 28520966 Free PMC article. Review.
Cited by
-
The Persistence of HIV Diversity, Transcription, and Nef Protein in Kaposi's Sarcoma Tumors during Antiretroviral Therapy.Viruses. 2022 Dec 13;14(12):2774. doi: 10.3390/v14122774. Viruses. 2022. PMID: 36560778 Free PMC article.
-
HIV persists throughout deep tissues with repopulation from multiple anatomical sources.J Clin Invest. 2020 Apr 1;130(4):1699-1712. doi: 10.1172/JCI134815. J Clin Invest. 2020. PMID: 31910162 Free PMC article.
-
Identification of Genetically Intact HIV-1 Proviruses in Specific CD4+ T Cells from Effectively Treated Participants.Cell Rep. 2017 Oct 17;21(3):813-822. doi: 10.1016/j.celrep.2017.09.081. Cell Rep. 2017. PMID: 29045846 Free PMC article.
-
The role of integration and clonal expansion in HIV infection: live long and prosper.Retrovirology. 2018 Oct 23;15(1):71. doi: 10.1186/s12977-018-0448-8. Retrovirology. 2018. PMID: 30352600 Free PMC article. Review.
-
Quantitation of the latent HIV-1 reservoir from the sequence diversity in viral outgrowth assays.Retrovirology. 2018 Jul 5;15(1):47. doi: 10.1186/s12977-018-0426-1. Retrovirology. 2018. PMID: 29976219 Free PMC article.
References
-
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278: 1295–300. - PubMed
-
- Wong JK, Hezareh M, Günthard HF, Havlir D V, Ignacio CC, Spina CA, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278: 1291–5. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

