ff14IDPs force field improving the conformation sampling of intrinsically disordered proteins
- PMID: 27484738
- PMCID: PMC5480619
- DOI: 10.1111/cbdd.12832
ff14IDPs force field improving the conformation sampling of intrinsically disordered proteins
Abstract
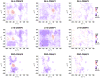
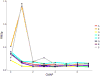
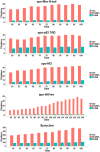
Intrinsically disordered proteins are proteins which lack of specific tertiary structure and unable to fold spontaneously without the partner binding. These intrinsically disordered proteins are found to associate with various diseases, such as diabetes, cancer, and neurodegenerative diseases. However, current widely used force fields, such as ff99SB, ff14SB, OPLS/AA, and Charmm27, are insufficient in sampling the conformational characters of intrinsically disordered proteins. In this study, the CMAP method was used to correct the φ/ψ distributions of disorder-promoting amino acids. The simulation results show that the force filed parameters (ff14IDPs) can improve the φ/ψ distributions of the disorder-promoting amino acids, with RMSD less than 0.10% relative to the benchmark data of intrinsically disordered proteins. Further test suggests that the calculated secondary chemical shifts under ff14IDPs are in quantitative agreement with the data of NMR experiment for five tested systems. In addition, the simulation results show that ff14IDPs can still be used to model structural proteins, such as tested lysozyme and ubiquitin, with better performance in coil regions than the original general Amber force field ff14SB. These findings confirm that the newly developed Amber ff14IDPs is a robust model for improving the conformation sampling of intrinsically disordered proteins.
Keywords: CMAP correction; IDPs; ff14IDPs; force field.
© 2016 John Wiley & Sons A/S.
Conflict of interest statement
There is no conflict of interest.
Figures
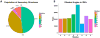


Similar articles
-
New force field on modeling intrinsically disordered proteins.Chem Biol Drug Des. 2014 Sep;84(3):253-69. doi: 10.1111/cbdd.12314. Epub 2014 Jul 1. Chem Biol Drug Des. 2014. PMID: 24589355
-
The IDP-Specific Force Field ff14IDPSFF Improves the Conformer Sampling of Intrinsically Disordered Proteins.J Chem Inf Model. 2017 May 22;57(5):1166-1178. doi: 10.1021/acs.jcim.7b00135. Epub 2017 May 4. J Chem Inf Model. 2017. PMID: 28448138 Free PMC article.
-
Intrinsically disordered protein-specific force field CHARMM36IDPSFF.Chem Biol Drug Des. 2018 Oct;92(4):1722-1735. doi: 10.1111/cbdd.13342. Epub 2018 Jul 1. Chem Biol Drug Des. 2018. PMID: 29808548
-
Conformational propensities of intrinsically disordered proteins from NMR chemical shifts.Chemphyschem. 2013 Sep 16;14(13):3034-45. doi: 10.1002/cphc.201300387. Epub 2013 Jun 21. Chemphyschem. 2013. PMID: 23794453 Review.
-
Recent Force Field Strategies for Intrinsically Disordered Proteins.J Chem Inf Model. 2021 Mar 22;61(3):1037-1047. doi: 10.1021/acs.jcim.0c01175. Epub 2021 Feb 16. J Chem Inf Model. 2021. PMID: 33591749 Free PMC article. Review.
Cited by
-
Assessment of transferable forcefields for protein simulations attests improved description of disordered states and secondary structure propensities, and hints at multi-protein systems as the next challenge for optimization.Comput Struct Biotechnol J. 2021 Apr 25;19:2626-2636. doi: 10.1016/j.csbj.2021.04.050. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34025949 Free PMC article.
-
A Practical Guide to All-Atom and Coarse-Grained Molecular Dynamics Simulations Using Amber and Gromacs: A Case Study of Disulfide-Bond Impact on the Intrinsically Disordered Amyloid Beta.Int J Mol Sci. 2024 Jun 18;25(12):6698. doi: 10.3390/ijms25126698. Int J Mol Sci. 2024. PMID: 38928405 Free PMC article.
-
Force Field Effects in Simulations of Flexible Peptides with Varying Polyproline II Propensity.J Chem Theory Comput. 2021 Oct 12;17(10):6634-6646. doi: 10.1021/acs.jctc.1c00408. Epub 2021 Sep 15. J Chem Theory Comput. 2021. PMID: 34524800 Free PMC article.
-
Conformational Dynamics of Intrinsically Disordered Proteins Regulate Biomolecular Condensate Chemistry.Chem Rev. 2022 Mar 23;122(6):6719-6748. doi: 10.1021/acs.chemrev.1c00774. Epub 2022 Feb 18. Chem Rev. 2022. PMID: 35179885 Free PMC article. Review.
-
Conformation Dynamics of the Intrinsically Disordered Protein c-Myb with the ff99IDPs Force Field.RSC Adv. 2017;7(47):29713-29721. doi: 10.1039/C7RA04133K. Epub 2017 Jun 7. RSC Adv. 2017. PMID: 29104751 Free PMC article.
References
-
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nature reviews Molecular cell biology. 2005;6:197–208. - PubMed
-
- Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Informatics Series. 2000:161–171. - PubMed
-
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. - PubMed
-
- Iakoucheva LM, Brown CJ, Lawson JD, Obradović Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. Journal of molecular biology. 2002;323:573–584. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

