A Prominent Site of Antibody Vulnerability on HIV Envelope Incorporates a Motif Associated with CCR5 Binding and Its Camouflaging Glycans
- PMID: 27438765
- PMCID: PMC4990068
- DOI: 10.1016/j.immuni.2016.06.026
A Prominent Site of Antibody Vulnerability on HIV Envelope Incorporates a Motif Associated with CCR5 Binding and Its Camouflaging Glycans
Abstract
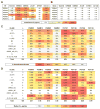
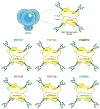
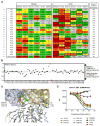
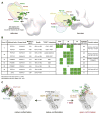
The dense patch of high-mannose-type glycans surrounding the N332 glycan on the HIV envelope glycoprotein (Env) is targeted by multiple broadly neutralizing antibodies (bnAbs). This region is relatively conserved, implying functional importance, the origins of which are not well understood. Here we describe the isolation of new bnAbs targeting this region. Examination of these and previously described antibodies to Env revealed that four different bnAb families targeted the (324)GDIR(327) peptide stretch at the base of the gp120 V3 loop and its nearby glycans. We found that this peptide stretch constitutes part of the CCR5 co-receptor binding site, with the high-mannose patch glycans serving to camouflage it from most antibodies. GDIR-glycan bnAbs, in contrast, bound both (324)GDIR(327) peptide residues and high-mannose patch glycans, which enabled broad reactivity against diverse HIV isolates. Thus, as for the CD4 binding site, bnAb effectiveness relies on circumventing the defenses of a critical functional region on Env.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Conserved Role of an N-Linked Glycan on the Surface Antigen of Human Immunodeficiency Virus Type 1 Modulating Virus Sensitivity to Broadly Neutralizing Antibodies against the Receptor and Coreceptor Binding Sites.J Virol. 2015 Oct 28;90(2):829-41. doi: 10.1128/JVI.02321-15. Print 2016 Jan 15. J Virol. 2015. PMID: 26512079 Free PMC article.
-
A Coreceptor-Mimetic Peptide Enhances the Potency of V3-Glycan Antibodies.J Virol. 2019 Feb 19;93(5):e01653-18. doi: 10.1128/JVI.01653-18. Print 2019 Mar 1. J Virol. 2019. PMID: 30541842 Free PMC article.
-
Glycoengineering HIV-1 Env creates 'supercharged' and 'hybrid' glycans to increase neutralizing antibody potency, breadth and saturation.PLoS Pathog. 2018 May 2;14(5):e1007024. doi: 10.1371/journal.ppat.1007024. eCollection 2018 May. PLoS Pathog. 2018. PMID: 29718999 Free PMC article.
-
Antibody responses to the HIV-1 envelope high mannose patch.Adv Immunol. 2019;143:11-73. doi: 10.1016/bs.ai.2019.08.002. Epub 2019 Sep 11. Adv Immunol. 2019. PMID: 31607367 Free PMC article. Review.
-
Broadly Neutralizing Antibodies for HIV Eradication.Curr HIV/AIDS Rep. 2016 Feb;13(1):31-7. doi: 10.1007/s11904-016-0299-7. Curr HIV/AIDS Rep. 2016. PMID: 26841901 Free PMC article. Review.
Cited by
-
Broadly neutralizing antibodies targeting HIV: Progress and challenges.Clin Immunol. 2023 Dec;257:109809. doi: 10.1016/j.clim.2023.109809. Epub 2023 Oct 16. Clin Immunol. 2023. PMID: 37852345 Free PMC article. Review.
-
Serum alpha-mannosidase as an additional barrier to eliciting oligomannose-specific HIV-1-neutralizing antibodies.Sci Rep. 2020 May 5;10(1):7582. doi: 10.1038/s41598-020-64500-8. Sci Rep. 2020. PMID: 32371950 Free PMC article.
-
Inhibiting the Ins and Outs of HIV Replication: Cell-Intrinsic Antiretroviral Restrictions at the Plasma Membrane.Front Immunol. 2018 Jan 4;8:1853. doi: 10.3389/fimmu.2017.01853. eCollection 2017. Front Immunol. 2018. PMID: 29354117 Free PMC article. Review.
-
Engineering immunogens that select for specific mutations in HIV broadly neutralizing antibodies.Nat Commun. 2024 Nov 3;15(1):9503. doi: 10.1038/s41467-024-53120-9. Nat Commun. 2024. PMID: 39489734 Free PMC article.
-
Beyond glycan barriers: non-cognate ligands and protein mimicry approaches to elicit broadly neutralizing antibodies for HIV-1.J Biomed Sci. 2024 Aug 21;31(1):83. doi: 10.1186/s12929-024-01073-y. J Biomed Sci. 2024. PMID: 39169357 Free PMC article. Review.
References
-
- Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol. 2011;85:9998–10009. - PMC - PubMed
-
- Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A, et al. Generation of human monoclonal-antibodies against HIV-1 proteins - electrofusion and Epstein-Barr virus transformation for peripheral-blood lymphocyte immortalization. AIDS Res Hum Retroviruses. 1994;10:359–369. - PubMed
-
- Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

