Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy
- PMID: 27432972
- PMCID: PMC4978246
- DOI: 10.1073/pnas.1609057113
Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy
Abstract
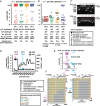
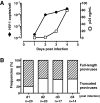
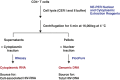
Despite years of plasma HIV-RNA levels <40 copies per milliliter during combination antiretroviral therapy (cART), the majority of HIV-infected patients exhibit persistent seropositivity to HIV-1 and evidence of immune activation. These patients also show persistence of proviruses of HIV-1 in circulating peripheral blood mononuclear cells. Many of these proviruses have been characterized as defective and thus thought to contribute little to HIV-1 pathogenesis. By combining 5'LTR-to-3'LTR single-genome amplification and direct amplicon sequencing, we have identified the presence of "defective" proviruses capable of transcribing novel unspliced HIV-RNA (usHIV-RNA) species in patients at all stages of HIV-1 infection. Although these novel usHIV-RNA transcripts had exon structures that were different from those of the known spliced HIV-RNA variants, they maintained translationally competent ORFs, involving elements of gag, pol, env, rev, and nef to encode a series of novel HIV-1 chimeric proteins. These novel usHIV-RNAs were detected in five of five patients, including four of four patients with prolonged viral suppression of HIV-RNA levels <40 copies per milliliter for more than 6 y. Our findings suggest that the persistent defective proviruses of HIV-1 are not "silent," but rather may contribute to HIV-1 pathogenesis by stimulating host-defense pathways that target foreign nucleic acids and proteins.
Keywords: HIV-1; activation; latency; provirus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
Defective HIV-1 proviruses produce viral proteins.Proc Natl Acad Sci U S A. 2020 Feb 18;117(7):3704-3710. doi: 10.1073/pnas.1917876117. Epub 2020 Feb 6. Proc Natl Acad Sci U S A. 2020. PMID: 32029589 Free PMC article.
-
Dynamic Shifts in the HIV Proviral Landscape During Long Term Combination Antiretroviral Therapy: Implications for Persistence and Control of HIV Infections.Viruses. 2020 Jan 25;12(2):136. doi: 10.3390/v12020136. Viruses. 2020. PMID: 31991737 Free PMC article.
-
The role of integration and clonal expansion in HIV infection: live long and prosper.Retrovirology. 2018 Oct 23;15(1):71. doi: 10.1186/s12977-018-0448-8. Retrovirology. 2018. PMID: 30352600 Free PMC article. Review.
-
Long-term persistence of transcriptionally active 'defective' HIV-1 proviruses: implications for persistent immune activation during antiretroviral therapy.AIDS. 2023 Nov 15;37(14):2119-2130. doi: 10.1097/QAD.0000000000003667. Epub 2023 Aug 22. AIDS. 2023. PMID: 37555786 Free PMC article.
-
Persistent HIV-1 transcription during ART: time to reassess its significance?Curr Opin HIV AIDS. 2024 May 1;19(3):124-132. doi: 10.1097/COH.0000000000000849. Epub 2024 Mar 12. Curr Opin HIV AIDS. 2024. PMID: 38502547 Free PMC article. Review.
Cited by
-
Intact proviral DNA assay analysis of large cohorts of people with HIV provides a benchmark for the frequency and composition of persistent proviral DNA.Proc Natl Acad Sci U S A. 2020 Aug 4;117(31):18692-18700. doi: 10.1073/pnas.2006816117. Epub 2020 Jul 20. Proc Natl Acad Sci U S A. 2020. PMID: 32690683 Free PMC article.
-
Phenotypic analysis of the unstimulated in vivo HIV CD4 T cell reservoir.Elife. 2020 Sep 29;9:e60933. doi: 10.7554/eLife.60933. Elife. 2020. PMID: 32990219 Free PMC article.
-
Structural and computational studies of HIV-1 RNA.RNA Biol. 2024 Jan;21(1):1-32. doi: 10.1080/15476286.2023.2289709. Epub 2023 Dec 15. RNA Biol. 2024. PMID: 38100535 Free PMC article. Review.
-
Despite early antiretroviral therapy effector memory and follicular helper CD4 T cells are major reservoirs in visceral lymphoid tissues of SIV-infected macaques.Mucosal Immunol. 2020 Jan;13(1):149-160. doi: 10.1038/s41385-019-0221-x. Epub 2019 Nov 13. Mucosal Immunol. 2020. PMID: 31723251 Free PMC article.
-
An Omics Approach to Extracellular Vesicles from HIV-1 Infected Cells.Cells. 2019 Jul 29;8(8):787. doi: 10.3390/cells8080787. Cells. 2019. PMID: 31362387 Free PMC article.
References
-
- DHHS Panel on Antiretoviral Guidelines for Adults and Adolescents 2015 Guidelines for the Use of Antiretroviral Agents in HIV-1-Infected Adults and Adolescents. Department of Health and Human Services. Available at https://aidsinfo.nih.gov/contentfiles/lvguidelines/adultandadolescentgl.pdf. Accessed July 1, 2016.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

