Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture
- PMID: 27432957
- PMCID: PMC4978254
- DOI: 10.1073/pnas.1609643113
Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture
Abstract
During interphase, the inactive X chromosome (Xi) is largely transcriptionally silent and adopts an unusual 3D configuration known as the "Barr body." Despite the importance of X chromosome inactivation, little is known about this 3D conformation. We recently showed that in humans the Xi chromosome exhibits three structural features, two of which are not shared by other chromosomes. First, like the chromosomes of many species, Xi forms compartments. Second, Xi is partitioned into two huge intervals, called "superdomains," such that pairs of loci in the same superdomain tend to colocalize. The boundary between the superdomains lies near DXZ4, a macrosatellite repeat whose Xi allele extensively binds the protein CCCTC-binding factor. Third, Xi exhibits extremely large loops, up to 77 megabases long, called "superloops." DXZ4 lies at the anchor of several superloops. Here, we combine 3D mapping, microscopy, and genome editing to study the structure of Xi, focusing on the role of DXZ4 We show that superloops and superdomains are conserved across eutherian mammals. By analyzing ligation events involving three or more loci, we demonstrate that DXZ4 and other superloop anchors tend to colocate simultaneously. Finally, we show that deleting DXZ4 on Xi leads to the disappearance of superdomains and superloops, changes in compartmentalization patterns, and changes in the distribution of chromatin marks. Thus, DXZ4 is essential for proper Xi packaging.
Keywords: CTCF; Hi‐C; X chromosome inactivation; genome engineering; inactive X chromosome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


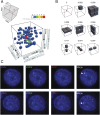
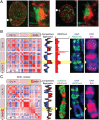
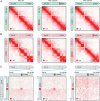
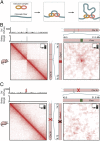
Similar articles
-
Characterization of the ICCE Repeat in Mammals Reveals an Evolutionary Relationship with the DXZ4 Macrosatellite through Conserved CTCF Binding Motifs.Genome Biol Evol. 2018 Sep 1;10(9):2190-2204. doi: 10.1093/gbe/evy176. Genome Biol Evol. 2018. PMID: 30102341 Free PMC article.
-
The macrosatellite DXZ4 mediates CTCF-dependent long-range intrachromosomal interactions on the human inactive X chromosome.Hum Mol Genet. 2012 Oct 15;21(20):4367-77. doi: 10.1093/hmg/dds270. Epub 2012 Jul 12. Hum Mol Genet. 2012. PMID: 22791747 Free PMC article.
-
Orientation-dependent Dxz4 contacts shape the 3D structure of the inactive X chromosome.Nat Commun. 2018 Apr 13;9(1):1445. doi: 10.1038/s41467-018-03694-y. Nat Commun. 2018. PMID: 29654302 Free PMC article.
-
Structural aspects of the inactive X chromosome.Philos Trans R Soc Lond B Biol Sci. 2017 Nov 5;372(1733):20160357. doi: 10.1098/rstb.2016.0357. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 28947656 Free PMC article. Review.
-
Multifaceted role of CTCF in X-chromosome inactivation.Chromosoma. 2024 Oct;133(4):217-231. doi: 10.1007/s00412-024-00826-w. Epub 2024 Oct 21. Chromosoma. 2024. PMID: 39433641 Review.
Cited by
-
ASHIC: hierarchical Bayesian modeling of diploid chromatin contacts and structures.Nucleic Acids Res. 2020 Dec 2;48(21):e123. doi: 10.1093/nar/gkaa872. Nucleic Acids Res. 2020. PMID: 33074315 Free PMC article.
-
Let-7, Lin28 and Hmga2 Expression in Ciliary Epithelium and Retinal Progenitor Cells.Invest Ophthalmol Vis Sci. 2021 Mar 1;62(3):31. doi: 10.1167/iovs.62.3.31. Invest Ophthalmol Vis Sci. 2021. PMID: 33749722 Free PMC article.
-
Analysis of differentially expressed genes on human X chromosome harboring large deletion induced by X-rays.J Radiat Res. 2023 Mar 23;64(2):300-303. doi: 10.1093/jrr/rrac093. J Radiat Res. 2023. PMID: 36617210 Free PMC article.
-
Xist exerts gene-specific silencing during XCI maintenance and impacts lineage-specific cell differentiation and proliferation during hematopoiesis.Nat Commun. 2022 Aug 1;13(1):4464. doi: 10.1038/s41467-022-32273-5. Nat Commun. 2022. PMID: 35915095 Free PMC article.
-
A TAD boundary is preserved upon deletion of the CTCF-rich Firre locus.Nat Commun. 2018 Apr 13;9(1):1444. doi: 10.1038/s41467-018-03614-0. Nat Commun. 2018. PMID: 29654311 Free PMC article.
References
-
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. - PubMed
-
- Brockdorff N, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71(3):515–526. - PubMed
-
- Brown CJ, et al. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71(3):527–542. - PubMed
-
- Dixon-McDougall T, Brown C. The making of a Barr body: The mosaic of factors that eXIST on the mammalian inactive X chromosome. Biochem Cell Biol. 2016;94(1):56–70. - PubMed
-
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434(7031):400–404. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

