Fragment Length of Circulating Tumor DNA
- PMID: 27428049
- PMCID: PMC4948782
- DOI: 10.1371/journal.pgen.1006162
Fragment Length of Circulating Tumor DNA
Abstract
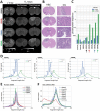
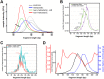
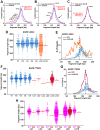
Malignant tumors shed DNA into the circulation. The transient half-life of circulating tumor DNA (ctDNA) may afford the opportunity to diagnose, monitor recurrence, and evaluate response to therapy solely through a non-invasive blood draw. However, detecting ctDNA against the normally occurring background of cell-free DNA derived from healthy cells has proven challenging, particularly in non-metastatic solid tumors. In this study, distinct differences in fragment length size between ctDNAs and normal cell-free DNA are defined. Human ctDNA in rat plasma derived from human glioblastoma multiforme stem-like cells in the rat brain and human hepatocellular carcinoma in the rat flank were found to have a shorter principal fragment length than the background rat cell-free DNA (134-144 bp vs. 167 bp, respectively). Subsequently, a similar shift in the fragment length of ctDNA in humans with melanoma and lung cancer was identified compared to healthy controls. Comparison of fragment lengths from cell-free DNA between a melanoma patient and healthy controls found that the BRAF V600E mutant allele occurred more commonly at a shorter fragment length than the fragment length of the wild-type allele (132-145 bp vs. 165 bp, respectively). Moreover, size-selecting for shorter cell-free DNA fragment lengths substantially increased the EGFR T790M mutant allele frequency in human lung cancer. These findings provide compelling evidence that experimental or bioinformatic isolation of a specific subset of fragment lengths from cell-free DNA may improve detection of ctDNA.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Quantitative analysis of the BRAF V600E mutation in circulating tumor-derived DNA in melanoma patients using competitive allele-specific TaqMan PCR.Int J Clin Oncol. 2016 Oct;21(5):981-988. doi: 10.1007/s10147-016-0976-y. Epub 2016 Apr 4. Int J Clin Oncol. 2016. PMID: 27041702
-
Detection of BRAF-V600E and V600K in melanoma circulating tumour cells by droplet digital PCR.Clin Biochem. 2015 Oct;48(15):999-1002. doi: 10.1016/j.clinbiochem.2014.12.007. Epub 2014 Dec 16. Clin Biochem. 2015. PMID: 25523300
-
Circulating tumor DNA to monitor treatment response and detect acquired resistance in patients with metastatic melanoma.Oncotarget. 2015 Dec 8;6(39):42008-18. doi: 10.18632/oncotarget.5788. Oncotarget. 2015. PMID: 26524482 Free PMC article.
-
The feasibility of using mutation detection in ctDNA to assess tumor dynamics.Int J Cancer. 2017 Jun 15;140(12):2642-2647. doi: 10.1002/ijc.30620. Epub 2017 Mar 2. Int J Cancer. 2017. PMID: 28124376 Free PMC article. Review.
-
Circulating tumour DNA (ctDNA) as a liquid biopsy for melanoma.Cancer Lett. 2017 Sep 28;404:62-69. doi: 10.1016/j.canlet.2017.06.030. Epub 2017 Jul 4. Cancer Lett. 2017. PMID: 28687355 Review.
Cited by
-
Biomarkers in Immunotherapy-Based Precision Treatments of Digestive System Tumors.Front Oncol. 2021 Mar 11;11:650481. doi: 10.3389/fonc.2021.650481. eCollection 2021. Front Oncol. 2021. PMID: 33777812 Free PMC article. Review.
-
Mining nucleic acid "omics" to boost liquid biopsy in cancer.Cell Rep Med. 2024 Sep 17;5(9):101736. doi: 10.1016/j.xcrm.2024.101736. Cell Rep Med. 2024. PMID: 39293399 Free PMC article. Review.
-
Prognostic and predictive significance of soluble programmed death ligand 1 in bronchoalveolar lavage fluid in stage IV non-small cell lung cancer.Transl Lung Cancer Res. 2024 Aug 31;13(8):1888-1906. doi: 10.21037/tlcr-24-392. Epub 2024 Aug 23. Transl Lung Cancer Res. 2024. PMID: 39263019 Free PMC article.
-
The Application of Liquid Biopsy Techniques in High-Risk Population for Hepatocellular Carcinoma.Cancer Manag Res. 2022 Sep 15;14:2735-2748. doi: 10.2147/CMAR.S373165. eCollection 2022. Cancer Manag Res. 2022. PMID: 36133739 Free PMC article. Review.
-
The cell-free DNA methylome captures distinctions between localized and metastatic prostate tumors.Nat Commun. 2022 Oct 29;13(1):6467. doi: 10.1038/s41467-022-34012-2. Nat Commun. 2022. PMID: 36309516 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

