Developmental regulation of myeloerythroid progenitor function by the Lin28b-let-7-Hmga2 axis
- PMID: 27401346
- PMCID: PMC4986532
- DOI: 10.1084/jem.20151912
Developmental regulation of myeloerythroid progenitor function by the Lin28b-let-7-Hmga2 axis
Abstract
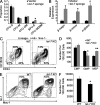
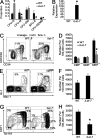
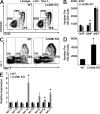
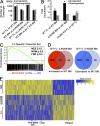
For appropriate development, tissue and organ system morphogenesis and maturation must occur in synchrony with the overall developmental requirements of the host. Mistiming of such developmental events often results in disease. The hematopoietic system matures from the fetal state, characterized by robust erythrocytic output that supports prenatal growth in the hypoxic intrauterine environment, to the postnatal state wherein granulocytes predominate to provide innate immunity. Regulation of the developmental timing of these myeloerythroid states is not well understood. In this study, we find that expression of the heterochronic factor Lin28b decreases in common myeloid progenitors during hematopoietic maturation to adulthood in mice. This decrease in Lin28b coincides with accumulation of mature let-7 microRNAs, whose biogenesis is regulated by Lin28 proteins. We find that inhibition of let-7 in the adult hematopoietic system recapitulates fetal erythroid-dominant hematopoiesis. Conversely, deletion of Lin28b or ectopic activation of let-7 microRNAs in the fetal state induces a shift toward adult-like myeloid-dominant output. Furthermore, we identify Hmga2 as an effector of this genetic switch. These studies provide the first detailed analysis of the roles of endogenous Lin28b and let-7 in the timing of hematopoietic states during development.
© 2016 Rowe et al.
Figures

Similar articles
-
The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells.Nat Cell Biol. 2013 Aug;15(8):916-25. doi: 10.1038/ncb2783. Epub 2013 Jun 30. Nat Cell Biol. 2013. PMID: 23811688
-
Distinct expression patterns predict differential roles of the miRNA-binding proteins, Lin28 and Lin28b, in the mouse testis: studies during postnatal development and in a model of hypogonadotropic hypogonadism.Endocrinology. 2013 Mar;154(3):1321-36. doi: 10.1210/en.2012-1745. Epub 2013 Jan 21. Endocrinology. 2013. PMID: 23337528
-
MYCN/LIN28B/Let-7/HMGA2 pathway implicated by meta-analysis of GWAS in suppression of post-natal proliferation thereby potentially contributing to aging.Mech Ageing Dev. 2013 Jul-Aug;134(7-8):346-8. doi: 10.1016/j.mad.2013.04.006. Epub 2013 Apr 29. Mech Ageing Dev. 2013. PMID: 23639551 Review.
-
TRIM71 suppresses tumorigenesis via modulation of Lin28B-let-7-HMGA2 signaling.Oncotarget. 2016 Nov 29;7(48):79854-79868. doi: 10.18632/oncotarget.13036. Oncotarget. 2016. PMID: 27821801 Free PMC article.
-
LIN28B and Let-7 in Diffuse Midline Glioma: A Review.Cancers (Basel). 2023 Jun 19;15(12):3241. doi: 10.3390/cancers15123241. Cancers (Basel). 2023. PMID: 37370851 Free PMC article. Review.
Cited by
-
The Zinc Finger Transcription Factor PLAGL2 Enhances Stem Cell Fate and Activates Expression of ASCL2 in Intestinal Epithelial Cells.Stem Cell Reports. 2018 Aug 14;11(2):410-424. doi: 10.1016/j.stemcr.2018.06.009. Epub 2018 Jul 12. Stem Cell Reports. 2018. PMID: 30017821 Free PMC article.
-
HMGA2 promotes long-term engraftment and myeloerythroid differentiation of human hematopoietic stem and progenitor cells.Blood Adv. 2019 Feb 26;3(4):681-691. doi: 10.1182/bloodadvances.2018023986. Blood Adv. 2019. PMID: 30808686 Free PMC article.
-
The Fetal-to-Adult Hematopoietic Stem Cell Transition and its Role in Childhood Hematopoietic Malignancies.Stem Cell Rev Rep. 2021 Dec;17(6):2059-2080. doi: 10.1007/s12015-021-10230-x. Epub 2021 Aug 23. Stem Cell Rev Rep. 2021. PMID: 34424480 Free PMC article. Review.
-
The efficiency of murine MLL-ENL-driven leukemia initiation changes with age and peaks during neonatal development.Blood Adv. 2019 Aug 13;3(15):2388-2399. doi: 10.1182/bloodadvances.2019000554. Blood Adv. 2019. PMID: 31405949 Free PMC article.
-
Cellular Basis of Embryonic Hematopoiesis and Its Implications in Prenatal Erythropoiesis.Int J Mol Sci. 2020 Dec 8;21(24):9346. doi: 10.3390/ijms21249346. Int J Mol Sci. 2020. PMID: 33302450 Free PMC article. Review.
References
-
- Bowie M.B., Kent D.G., Dykstra B., McKnight K.D., McCaffrey L., Hoodless P.A., and Eaves C.J.. 2007. Identification of a new intrinsically timed developmental checkpoint that reprograms key hematopoietic stem cell properties. Proc. Natl. Acad. Sci. USA. 104:5878–5882. 10.1073/pnas.0700460104 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

